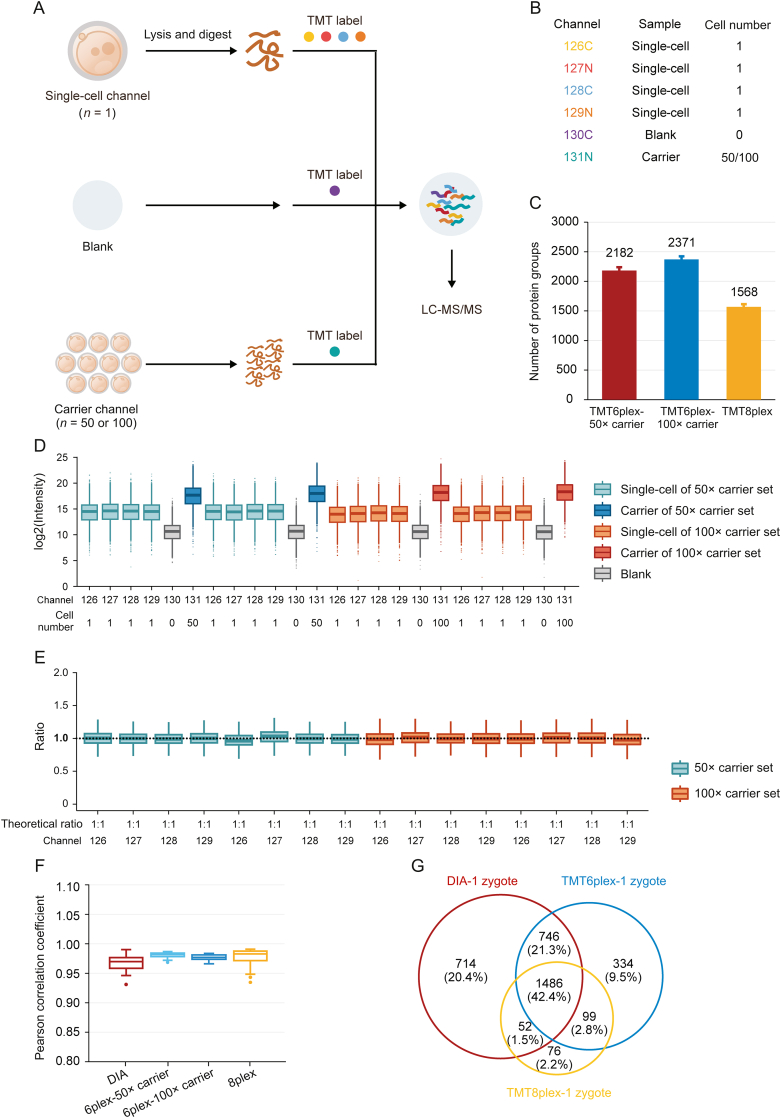
Fig. 3.
High-throughput proteomic profiling of mouse zygotes by single-cell proteomics. (A) Workflow for characterizing the mouse zygote proteome by high-throughput single-cell proteomics with the carrier channel. (B) Channel design for TMT6plex single-cell proteomics. Each TMT6plex experiment included one blank channel (0 zygote), one carrier channel (50 or 100 oocytes), and four single-cell channels (1 zygote for each). (C) Comparison of the number of protein groups identified by multiplexed single-cell proteomics with TMT6plex-50× carrier, TMT6plex-100× carrier, and TMT8plex. Data in the plot are presented as mean ± standard deviation. (D) Intensity distribution of all proteins in each TMT channel. Each different carrier TMT6plex experiment had two replicates. Boxplots show the median (middle bar), 25th and 75th percentiles (box), and 1.5× interquartile range (whiskers). (E) Boxplots of measuring ratios of protein groups based on tandem mass tags (TMT) intensities between different single-cell channels in each TMT6plex experiment. The denominator is the quantitative mean of 16 single-cell channels in four TMT experiments. Boxplots show the median (middle bar), 25th and 75th percentiles (box), and 1.5× interquartile range (whiskers). (F) Comparison of Pearson's correlation coefficients between each run (or single-cell channel) of data-independent acquisition (DIA) low-input proteomics, TMT6plex-50× carrier, TMT6plex-100× carrier, and TMT8plex single-cell proteomics. Boxplots show the median (middle bar), 25th and 75th percentiles (box), and 1.5× interquartile range (whiskers). (G) Venn diagram shows the overlap between DIA low-input proteomics, TMT6plex, and TMT8plex single-cell proteomics identified proteins from single mouse zygotes. LC-MS/MS: liquid chromatography tandem mass spectrometry.