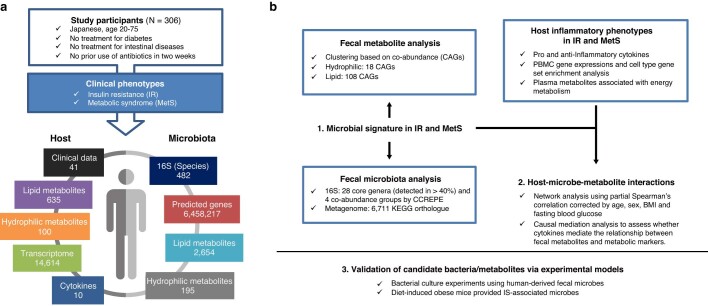
Extended Data Fig. 1. Overview of multi-omics analysis and data.
a, Individuals without a prior diagnosis of diabetes, diabetic medications, or intestinal diseases were included (n = 306). Insulin resistance (IR) and metabolic syndrome (MetS) were the main clinical phenotypes. To evaluate the host-microbe relationship, we collected 1) host factors: clinical, plasma metabolome, peripheral blood mononuclear cells (PBMC) transcriptome, and cytokine data, and 2) microbial factors: 16S rRNA pyrosequencing, shotgun metagenome, and faecal metabolome. The numbers of elements after quality filtering are shown for each data set. b, The multi-omics analysis workflow. To identify the microbes that affect metabolic phenotypes, we first analysed the phenotype-associated metabolomic signatures by binning metabolites into co-abundance groups (CAGs). Microbial signatures were determined using the 16S and metagenomic datasets, and their associations with metabolites were analysed. To gain insight into the host-microbe relationship, the associations among faecal metabolites/microbes and host plasma metabolites, cytokines, and PBMC genes were analysed. We also assessed the mediation effects of plasma cytokines on the relationships between faecal metabolites and metabolic markers. Finally, to validate the effects of candidate metabolites/microbes on metabolic phenotypes, we performed bacterial culture and animal experiments. The associations between clinical phenotypes and omics markers were adjusted by age and sex wherever appropriate.