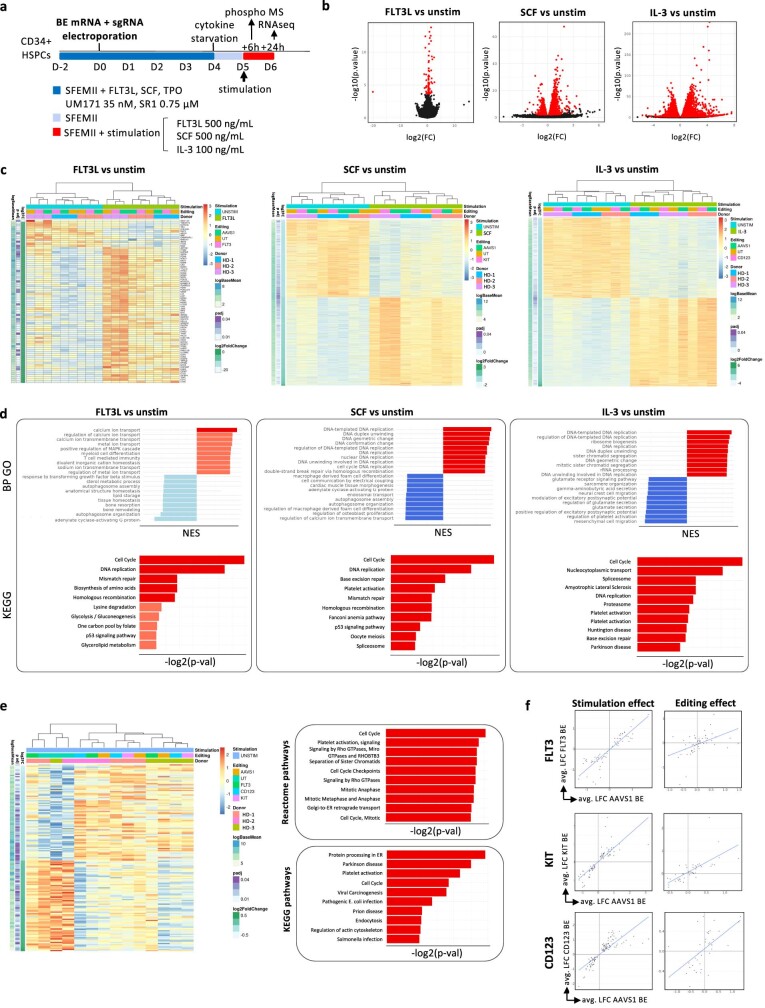
Extended Data Fig. 5. Epitope editing does not affect downstream signalling and transcriptional response of CD34+ HSPCs upon ligand stimulation.
a. Experimental layout for RNAseq and phospho-profiling by mass spectrometry of epitope edited CD34+ HSPCs. Cells were thawed and edited as previously described, cultured for 4 days to allow substitution of the receptor protein on the cell surface, starved for cytokines (24h) and stimulated with the respective ligand (FLT3L, IL-3, SCF). Cells were harvested at 6h and 24h after the start of stimulation for phospho-MS and RNAseq respectively. The experiments were performed in biological replicate (N = 3 and N = 2 CD34+ donors for RNAseq and MS, respectively). b. Volcano plots showing significantly differentially expressed genes (DEGs) in response to each receptor stimulation. The full list is available in Supplementary Tables 3 to 5. c. Heatmaps reporting the significant DEGs for each receptor/ligand couple (FDR-corrected p<0.05). Each column corresponds to a sample (see mapping reported above the heatmap and legend on the right), while rows correspond to gene transcripts. As depicted by the dendrogram, unbiased clustering shows predominant effects of the (1) stimulation and (2) donor, while editing conditions are distributed within each cluster. Full comparisons are available in Supplementary Tables 10–12. d. Enrichment analyses for the comparison of (from left to right) FLT3L, IL-3 and SCF vs unstimulated. Top row reports the GSEA analysis for the Biological Process (BP) gene ontology. The top 10 terms by significance for NES<0 or NES>0 are reported. Dark red, FDR<0.05 and NES>0, Light red FDR>0.05 and NES>0, Dark blue FDR<0.05 and NES<0, Light blue FDR>0.05 and NES<0. The bottom row reports top 10 terms by significance for KEGG pathways enrichment analysis. Dark red, FDR < 0.05, Light Red FDR > 0.05. The full list of terms is available in Supplementary Tables 7–9. e. Left, Heatmap of differentially expressed genes (DEGs) from the UT vs AAVS1 comparison (editing effect) on CD34+ HSPC RNAseq samples. Full list of DEGs is available in Supplementary Table 6. Right, enrichment analysis of the UT vs AAVS1 DEGS, the top 10 terms by significance for KEGG and Reactome pathways are reported. Full lists are available in Supplementary Tables 7–9. f. Phospho-proteomic analysis of edited CD34+ HSPCs by mass spectrometry. Scatter plot showing the concordance of the log Fold Change (LFC) of phosphorylated sites by MS upon stimulation in receptor edited vs AAVS1 control condition. Sites differentially with differential phosphorylation associated with the Stimulation (left column) or the Editing (right column) are reported in the scatter plots. Heatmaps reporting the phospho-sites for each comparison and editing condition are reported in Supplementary Fig. 3. Full lists of phospho-sites for each comparison are available as Supplementary Tables 13–15.