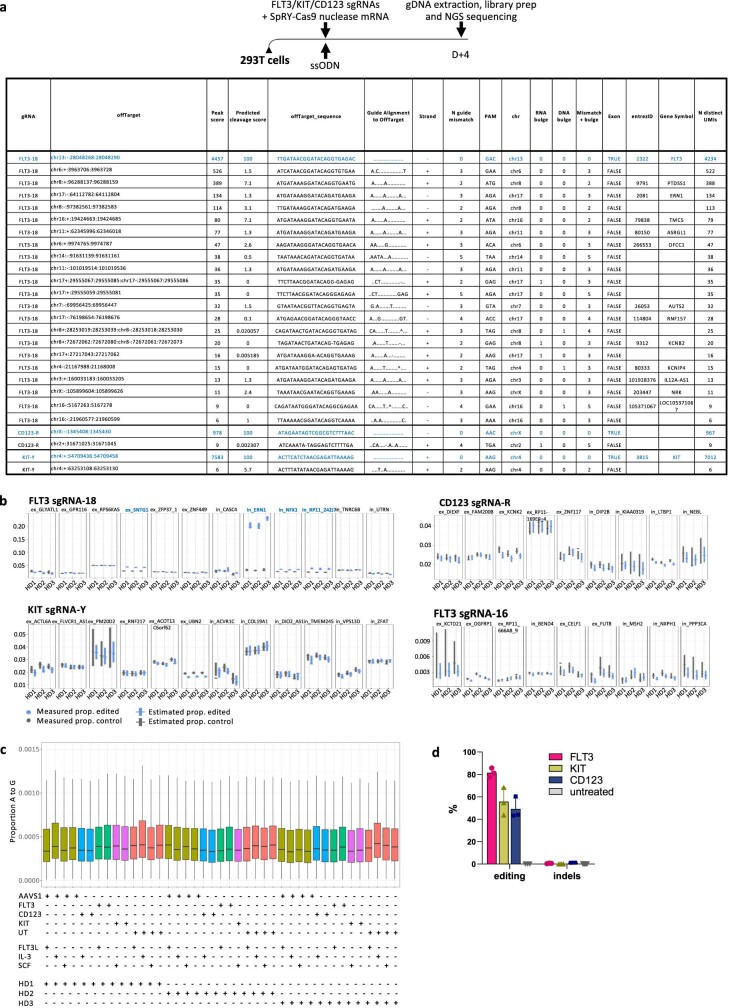
Extended Data Fig. 8. Off-target analyses of FLT3, CD123 and KIT epitope editing.
a GUIDE-Seq experimental design on 293T cells. GUIDE-Seq results were filtered by number of gRNA mismatches+bulge <6. The detected on-target locus is highlighted in blue. b Estimation plots of off-target deamination at predicted sites by NGS sequencing for FLT3-sgRNA-18, CD123-sgRNA-R, KIT-sgRNA-Y and FLT3-sgRNA-16. Loci with significant excess deamination compared to control are highlighted in blue. Additional information on predicted OT sites is reported in Supplementary Fig. 3 and Supplementary Tables 17–20. c Random deamination within RNAseq data of edited CD34+ HSPCs. The proportion of A-to-G conversion observed on the top 5% reads by coverage on RNAseq samples of edited CD34+ HSPCs is reported. Sample editing condition, whether it was stimulated with each ligand and donor source are reported below. Threshold and coverage are reported in Supplementary Table 21. d. On-target editing efficiency and indel formation in CD34+ HSPCs (%). Indel frequency was calculated on NGS samples as the proportion of reads harbouring any deletion or base insertion within the sgRNA sequence.