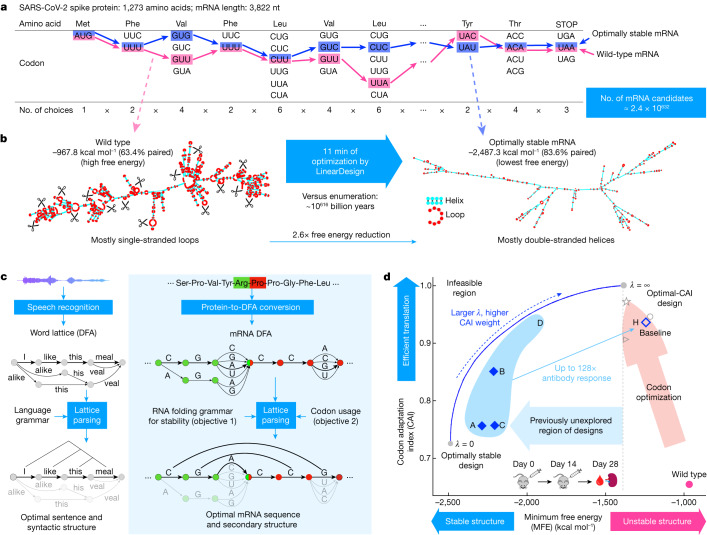
Fig. 1. Overview of mRNA coding region design for both stability and codon optimality using SARS-CoV-2 spike protein as an example.
a, Due to codon degeneracy and combinatorial explosion, there are around 2.4 × 10632 possible mRNA sequences encoding the spike protein. Enumerating every possible sequence would take around 10616 billion years. The pink and blue paths represent the wild-type and the optimally stable (lowest free energy) sequences, respectively. nt, nucleotides. b, The secondary structures of wild-type (left) and optimally stable (right) spike mRNAs. The wild-type mRNA is mostly single-stranded and thus prone to degradation in loop regions (red), whereas the optimally stable mRNA is mostly double-stranded. Optimization using LinearDesign takes around 11 min. c, The application of DFA and lattice parsing in computational linguistics (left) and its adaptation to mRNA design (right). An mRNA DFA (analogous to a word lattice) compactly encodes all mRNA candidates, which are folded simultaneously by lattice parsing to find the optimal mRNA (Fig. 2). d, Two-dimensional visualization of the mRNA design space, with stability (represented by MFE) on the x axis and codon optimality (represented by CAI) on the y axis. The standard mRNA design method of codon optimization improves codon usage (pink arrow) but is unable to explore the high-stability region (left of the dashed line); this standard approach is exemplified by the COVID-19 mRNA vaccine products BNT-162b2 (BioNTech-Pfizer, circle), mRNA-1273 (Moderna, star) and CVnCoV/CV2CoV (CureVac, wedge). LinearDesign jointly optimizes stability and codon optimality (blue curve, with λ being the weight assigned to codon optimality). We selected seven mRNA designs (four (A–D) are shown here) and a codon-optimized baseline (H) for in vitro and in vivo experiments (Fig. 4).