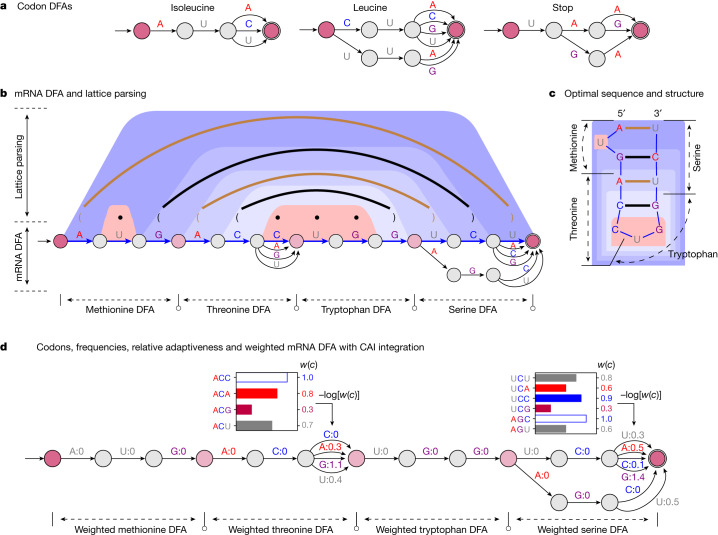
Fig. 2. Illustration of the LinearDesign algorithm.
a, Codon DFAs. b, An mRNA DFA (bottom) and lattice parsing on that DFA (top). In the DFA, the optimal mRNA sequence under a simplified energy model is shown as the blue path, together with its optimal structure shown in the dot-bracket format (dots indicate unpaired, and brackets indicate base pairs). In lattice parsing, the brown and black arcs also depict base pairs (two GC pairs and two AU pairs), and the trapezoidal shaded areas depict the decomposition of the optimal structure. Among all mRNA sequences encoded in the DFA, lattice parsing finds the optimal sequence with its optimal structure, achieving the lowest free energy under this energy model, where GC and AU pairs have energies of −3 and −2 kcal mol−1, respectively (Extended Data Fig. 1e). Note that here we use the simplified energy model for illustration, but our implementation uses the nearest-neighbour energy model. c, Another illustration of the optimal sequence and secondary structure in b. d, Joint optimization between stability and codon optimality by integrating the codon optimality in weighted DFAs. Top, the codon frequencies for threonine and serine. The relative adaptiveness w(c) of a codon c is the ratio of the frequency of c to the frequency of the most frequent codon encoding the same amino acid (white bar), and its value is shown to the right of the bars. Bottom, a weighted mRNA DFA encodes the CAI of each candidate in the total weight of its corresponding path by using –log[w(c)] (the cost of choosing codon c) as edge weights (Methods, ‘Optimization objectives’). This weighted DFA is used as input to lattice parsing for joint optimization between stability and codon optimality.