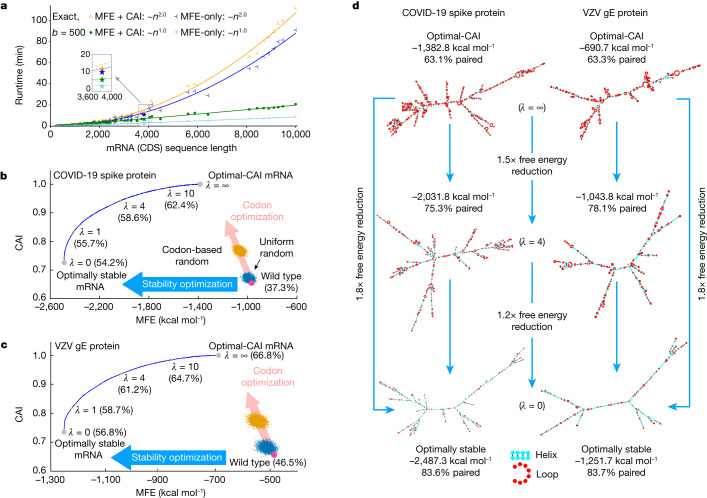
Fig. 3. Computational characteristics of the LinearDesign algorithm.
a, Runtime analysis of mRNA design for proteins in UniProt (Supplementary Table 1). Overall, our exact search scales quadratically with sequence length (Supplementary Figs. 7 and 8), and our MFE + CAI mode (with λ = 4) is about 15% slower than the MFE-only version. Moreover, beam search (b = 500) significantly speeds up the design of long sequences, with minor search errors (Supplementary Fig. 9). b,c, Two-dimensional (MFE–CAI) visualizations of designs for the SARS-CoV-2 spike (b) and VZV gE (c) proteins, respectively (both using human codon preference). The blue curves form the feasibility limit (optimal boundary), by varying λ from 0 to ∞ (see Extended Data Fig. 4 for λ of (–∞, 0]). The GC percentage is shown in parentheses. The human genome favours GC-rich codons; therefore, codon optimization (pink arrows) also improves stability, but only marginally, as the two optimization directions (codon versus stability) are largely orthogonal. By contrast, with an AU-rich codon preference (such as in yeast), codon optimization decreases stability (Extended Data Fig. 4b). d, Secondary structures of the mRNA designs for SARS-CoV-2 spike and VZV gE protein. The optimal-CAI designs (top, λ = ∞) are largely single-stranded (around 60% base-paired), whereas the optimally stable designs (bottom, λ = 0) are mostly double-stranded (around 80% base-paired). We also show intermediate designs (centre, λ = 4) with a balance of stability and CAI.