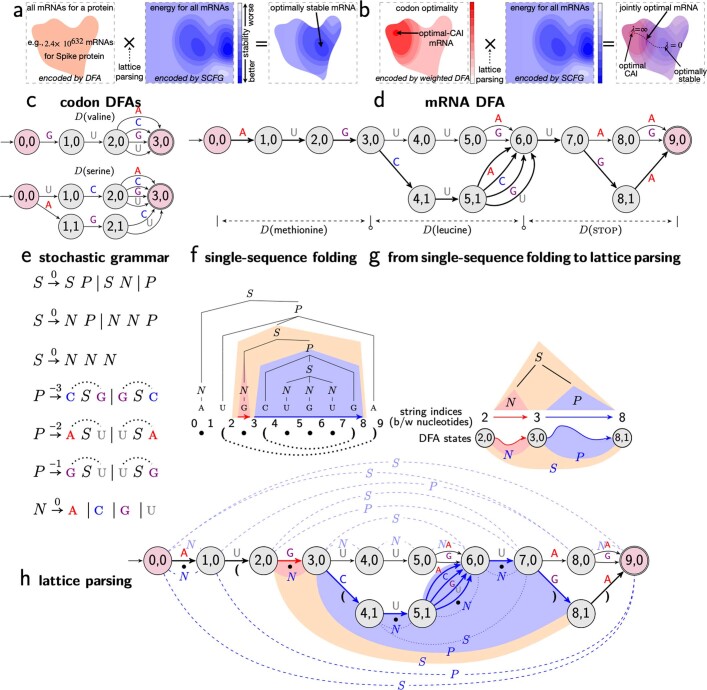
Extended Data Fig. 1. Illustrations of the optimization problems in mRNA design, DFA representations, single sequence folding as natural language parsing, and lattice parsing.
a–b, Visualization of mRNA design as optimization problems for stability (objective 1, in a) and joint stability and codon optimality (objectives 1 and 2, in b). c–h show how lattice parsing solves the first optimization problem (see Fig. 2d for the second). c, Codon DFAs. d, An mRNA DFA made of three codon DFAs. The thick paths depict the optimal mRNA sequences under the simplified energy model in e, AUGCU⋆UGA, where ⋆ could be any nucleotide. e, Stochastic context-free grammar (SCFG) for a simplified folding free energy model. Each rule has a cost (i.e., energy term, the lower the better), and the dotted arcs represent base pairs in RNA secondary structure. f, Single-sequence folding is equivalent to context-free parsing with an SCFG; the parse tree represents the best secondary structure for the input mRNA sequence. g, We extend single-sequence parsing (top) to lattice parsing (bottom) by replacing the input string with a DFA, where each string index becomes a DFA state, and a span becomes a path between two states. h, Lattice parsing with the grammar in e for the DFA in d. The blue arcs below the DFA depict the (shared) best structure for the optimal sequences AUGCU⋆UGA in the whole DFA, while the dashed light-blue arcs above the DFA represent the best structure for a suboptimal sequence AUGUUAUAA. Lattice parsing can also incorporate codon optimality (objective 2, see b), by replacing the DFA with a weighted one (Fig. 2d).