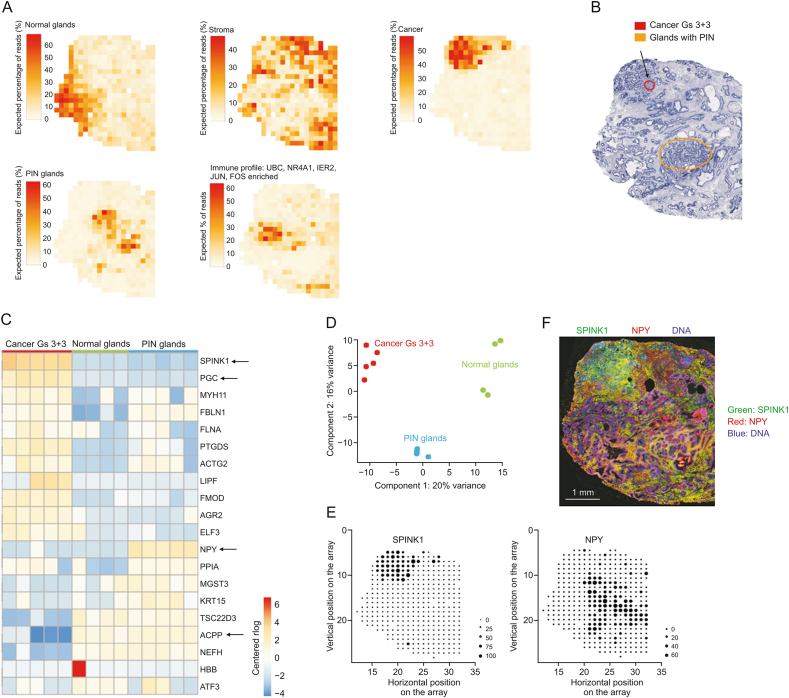
Fig. 4.
Spatial gene expression heterogeneity within the cancer tissue sample. (A) Factor activity maps for selected factors corresponding to epithelial, stromal, cancerous, prostatic intraepithelial neoplasia (PIN), or inflamed regions. (B) Annotated brightfield image of hematoxylin and eosin (H&E)-stained tissue section. (C) Heatmap of 20 most variable genes between cancer, PIN, and normal gland regions. Arrows highlight genes of interest validated by immunohistochemistry (IHC). (D) First two principal components of spot sets from C separate cancer, PIN, and normal regions. (E) Array dot plots for serine peptidase inhibitor kazal type 1 (SPINK1) and NPY. Circle size in array dot plots indicates normalized spatial transcriptomics (ST) counts. (F) IHC staining for SPINK1 and neuropeptide Y (NPY) of an adjacent section on the ST array. Nuclei are stained with 4',6-diamidino-2-phenylindole (DAPI; blue). Scale bar indicates 1 mm. UBC: ubiquitin C; NR4A1: nuclear receptor subfamily 4 group A member 1; IER2: immediate early response protein 2. Reprinted from Ref. [112] with permission.