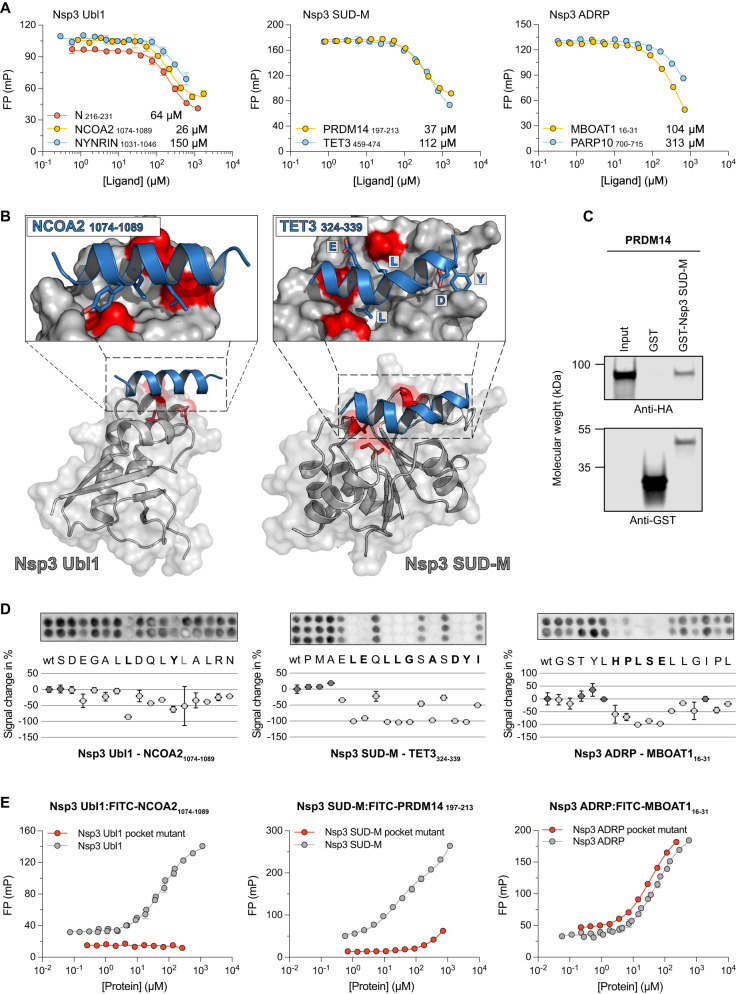
Fig. 3. Biophysical analysis of the interactions between Nsp3 Ubl1, Nsp3 ADRP, and Nsp3 SUD-M, with peptide ligands from human proteins.
A Fluorescence polarization-monitored displacement experiments measuring the affinity between globular domains of Nsp3 and peptide ligands from disordered regions of the human proteome identified by phage display. The data are presented as means ± SD (N = 3). The KD values for these and all subsequent affinity measurements performed in this study are shown next to each peptide and in Supplementary Data 4. B ColabFold structural predictions for the interaction between the globular domains of Nsp3 and the peptide ligands. The globular domains of Nsp3 are shown in gray, whereas the peptides are shown as blue ribbons. The residues that were mutated for the binding pocket analysis are highlighted in red (Ubl1: V852K, L889S, and L893D; SUD-M: A1397E, V1453K, S1478D) and the residues that were identified to be important for binding by SPOT arrays are shown as sticks. In the case of Nsp3 SUD-M the N terminus of the helix is on the left side of the enlarged panel. The ColabFold pLDDT confidence scores were high for the globular domains of Nsp3 (>90) but varied widely for peptide predictions (>80 for interactions with Nsp3 Ub1 and 40–60 for Nsp3 SUD-M) and are shown in Supplementary Fig. 4. C Pulldown of full length PRDM14 by GST-tagged Nsp3 SUD-M as visualized by Western blot. Molecular weight is indicated. Original blots for these and all subsequent Western blot experiments are provided in the Source Data file (repeated in two independent experiments). D SPOT array alanine scans for the indicated peptides. Residues involved in binding are shown in bold. Signal intensities were normalized to wild type (wt) and presented as average percent signal change. Error bars indicate one standard deviation from the average (mean ± SD; N ≥ 2). E Fluorescence polarization-monitored saturation experiments measuring the affinity between labeled peptides and globular domains of Nsp3 or the pocket mutant variants of them. The data are presented as means ± SD (N = 6 for Nsp3 Ubl1 and Nsp3 ADRP measurements and N = 3 for all other).