Extended Data Fig. 4. EGR2 does not control chromatin accessibility in TH17 cells.
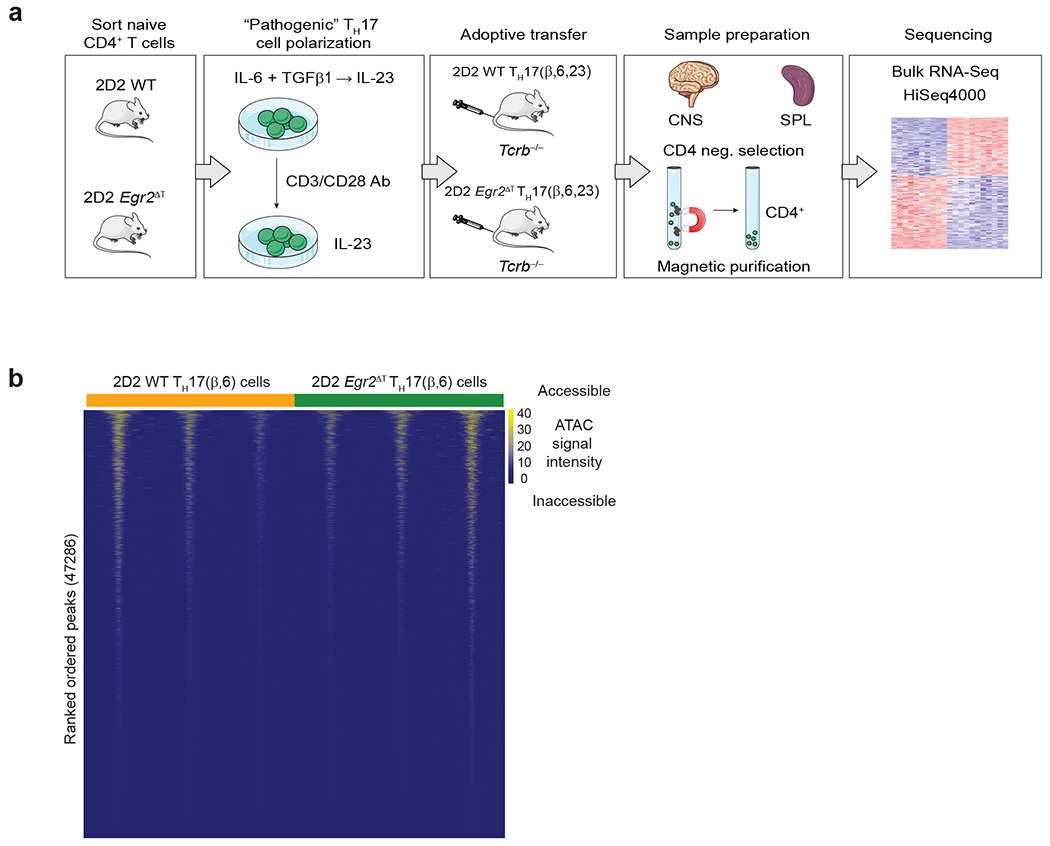
a, Bulk RNA-seq Workflow. Naïve 2D2 WT and 2D2 Egr2ΔT CD4+ T cells were activated and differentiated under pathogenic TH17 cell-polarizing conditions (TH17(β,6,23)) for 5 days. Differentiated TH17(β,6,23) cells were reactivated by plate-bound CD3+CD28 antibodies in the presence of IL-23 for 48h before adoptive transfer into Tcrb−/− recipients. At the peak of the disease (day 20 post-transfer), donor CD4+ T cells were purified from spleens and CNS of the Tcrb−/− recipient mice using CD4 negative selection for RNA profiling and library was sequenced on a HiSeq4000. Three independent experiments were performed. b, Heatmap illustrating dynamics of chromatin accessibility in 2D2 WT and 2D2 Egr2ΔT TH17(β,6) cells (GSE224960). Data represent n = 3 biologically independent replicates per condition.
