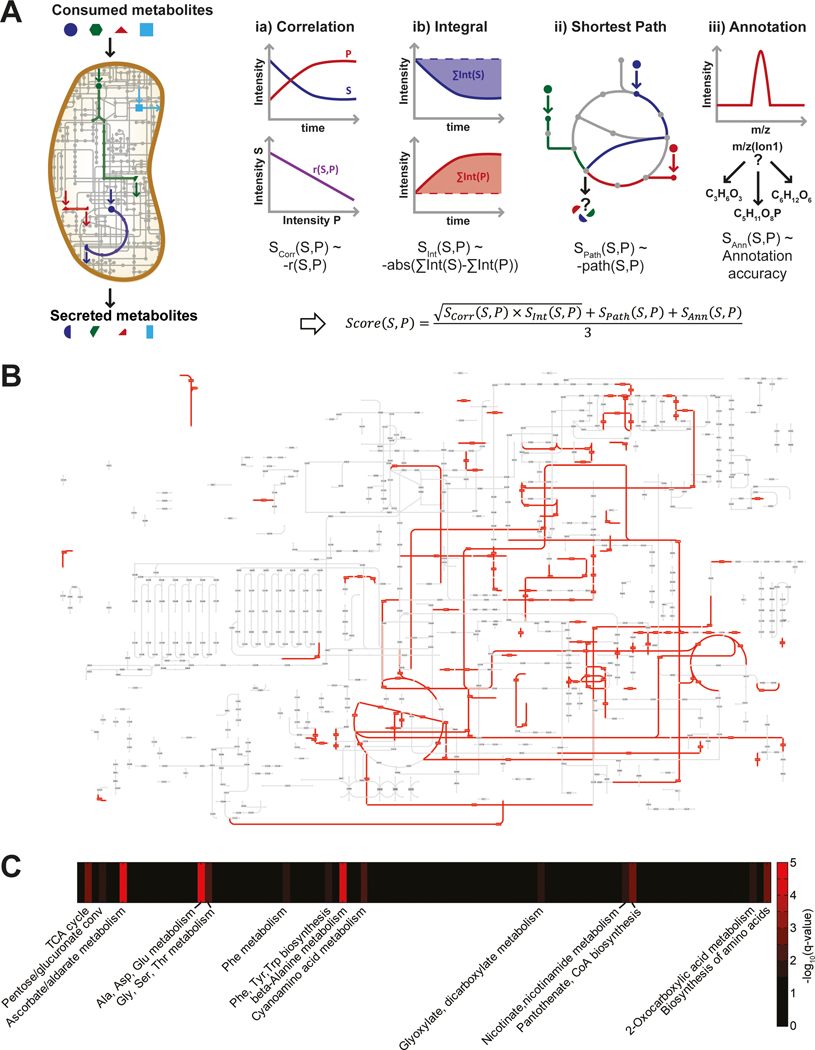
Fig. 2. Identification of the most probable carbon fluxes using a genome-scale metabolic model.
(A) Scheme of the algorithm scoring metabolic pathways converting consumed substrates into secreted metabolites. (B) KEGG metabolic pathway map overlaid with the 250 top pathway hits showing a coverage of 12.1%. (C) Enrichment analysis of the top one percent hits for their metabolic pathway definition.