Abstract
Evidence suggests that shared pathophysiological mechanisms in neuropsychiatric disorders (NPDs) may contribute to risk and resilience. We used single-gene and network-level transcriptomic approaches to investigate shared and disorder-specific processes underlying posttraumatic stress disorder (PTSD), Parkinson’s disease (PD) and schizophrenia in a South African sample. RNA-seq was performed on blood obtained from cases and controls from each cohort. Gene expression and weighted gene correlation network analyses (WGCNA) were performed using DESeq2 and CEMiTool, respectively. Significant differences in gene expression were limited to the PTSD cohort. However, WGCNA implicated, amongst others, ribosomal expression, inflammation and ubiquitination as key players in the NPDs under investigation. Differential expression in ribosomal-related pathways was observed in the PTSD and PD cohorts, and focal adhesion and extracellular matrix pathways were implicated in PD and schizophrenia. We propose that, despite different phenotypic presentations, core transdiagnostic mechanisms may play important roles in the molecular aetiology of NPDs.
Supplementary Information
The online version contains supplementary material available at 10.1007/s44192-022-00009-y.
Introduction
Psychiatric disorders, such as posttraumatic stress disorder (PTSD) and schizophrenia, and neurodegenerative disorders, such as Parkinson’s disease (PD), pose an immense burden to society [1, 2]. PTSD is a stress-related disorder that occurs consequent to an identifiable proximate traumatic event. It has a prevalence of approximately 8% in the general population [3]. The disorder is particularly relevant in South Africa, given the high trauma burden, as reported by the South African Stress and Health Study [4]. Schizophrenia is a severe psychiatric disorder that affects almost 1% of the world’s population [5]. The illness typically has an onset in early adulthood and patients frequently experience multiple psychotic relapses and admissions to hospital, enduring deficits in cognitive, social and occupational functioning, and a significantly decreased life expectancy [6–8]. PD is a progressive, debilitating disease with substantial physical, psychological and social implications, and is the second most common neurodegenerative disorder after Alzheimer’s disease (AD). The global age-standardised rate for PD is 33 per 100,000 people, adjusted for disability life-years or DALYs [9].
PTSD, schizophrenia and PD are all multifactorial disorders, with a combination of genetic and environmental factors contributing substantially to their aetiology [10–12]. However, despite recent advances in biomarker and genetics studies, the pathophysiology of both PTSD and schizophrenia remains poorly understood. Likewise, although pathologically characterised by loss of nigrostriatal dopaminergic innervation, PD is considered an idiopathic disorder with Mendelian inheritance occurring in only 3−5% of patients [13].
Shared aetiologies may underlie these neuropsychiatric disorders (NPDs). For example, in a recent genome-wide association study (GWAS), it was found that PRKN, a PD-associated gene involved in proteasome-dependent degradation of proteins, was found to be associated with PTSD in a European population [14]. This finding was supported recently by research from our group [15]. Indeed, accumulating evidence indicates that PTSD is associated with increased risk for neurodegenerative disorders, such as PD and AD [16–18]. In addition, individuals who develop adjustment disorder as a result of inability to cope with stress have been found to be more likely to develop PD later in life [19]. Studies have also shown that stress can induce neurotoxicity in brain regions such as the substantia nigra, prefrontal cortex and hippocampus, resulting in neurodegenerative pathological alterations [20].
The association between schizophrenia and PD is evident on a clinical as well as molecular level. Parkinson-like motor abnormalities, independent of antipsychotic treatment, have been observed in patients with schizophrenia spectrum disorders [21–24] and PD patients often exhibit pathological features that overlap with schizophrenia, including hallucinations, psychosis and cognitive dysfunction [25–28]. In addition, a recent GWAS investigation identified nine loci that were jointly associated with both PD and schizophrenia [29], with seven of these loci common in previous studies [30–34]. Deletions in 22q11 have been found to be a strong genetic risk factor for schizophrenia [35] and a number of studies suggest that individuals with 22q11 deletions also develop early-onset PD [36–39]. On a molecular level, alterations in dopaminergic neurotransmission play a central role in both disorders—PD is associated with loss of dopaminergic neurons of the substantia nigra pars compacta, whereas schizophrenia has been found to be associated with hypodopaminergic activity in the prefrontal cortex and hyperactive dopaminergic activity in the mesolimbic and striatal regions of the brain [40, 41].
Prevalence estimates of PTSD in schizophrenia range from 12 to 29%, which is higher than that for the general population [42–44]. Psychotic symptoms typical of schizophrenia have been found to occur with a higher frequency than expected in PTSD patients [45–47]. In addition, symptoms of depersonalisation and derealisation, which form a dissociative subtype of PTSD, are also exhibited by patients with schizophrenia [48]. Although there is limited research into the genetic overlap between the two disorders, polygenic risk score analysis indicates a genetic overlap between PTSD and schizophrenia [49].
Due to converging lines of genetic, molecular, neurobiological and phenotypic evidence supporting shared pathophysiology underlying PTSD, PD and schizophrenia, we aimed to identify unique as well as shared pathways underlying these three disorders. We employed RNA-seq whole-transcriptome analysis, followed by analysis of genes that are co-expressed within modular networks identified within each disease cohort, using weighted gene correlation network analysis (WGCNA). WGCNA is a co-expression analysis that clusters genes with highly correlated expression levels into numerous modules, each module comprising genes with similar expression patterns. This facilitates our understanding of how genes within the modules may interact with one another, and may shed light on biological mechanisms underlying the disorders in question, as genes with similar function will usually exhibit strong correlations in expression [50, 51]. Highly-connected genes within modules, known as “hub genes”, are often important to the functionality of the module [52], and as such, may play a role in the aetiology of the disorder. Compared to analyses at the single-gene level, WGCNA represents an integrated, network-level approach, enabling the identification of altered gene co-expression within modules, and permits identification of broad pathological mechanisms underlying PTSD, PD and schizophrenia, even if the contributing genes are disorder-specific.
Methods
The three diagnostic cohorts were recruited as part of the Shared Roots Study, conducted in Cape Town, South Africa. The study was approved by Stellenbosch University’s Health Research Ethics Committee (Ethics Approval Number: N13/08/115). Participants were recruited through purposive sampling at Stellenbosch University Faculty of Medicine and Health Sciences (Tygerberg Campus) and at Stikland Hospital, in Cape Town. Inclusion criteria for all participants was that they were willing and able to provide informed consent; were 18 years and older; all self-identified as being part of the South African Coloured population (as termed in the South African census); and were able to read and write Afrikaans or English. Participants were excluded if they did not provide informed consent; were pregnant or were younger than 18 years of age.
Demographic and clinical assessment
A sociodemographic questionnaire was administered to gather data including age, sex, education, employment, income, marital status and ethnicity. A participant was considered a smoker if he/she had ever smoked cigarettes in their lifetime. Likewise, a participant was considered to have consumed alcohol if he/she had done so in their lifetime. We did not quantify alcohol consumption for this analysis.
The main criterion separating cases and controls in each of the cohorts rested on the clinical diagnosis of the disorder under investigation in each cohort–PTSD, schizophrenia or Parkinson’s Disease. All other inclusion and exclusion criteria are the same for cases and controls within each cohort.
Diagnosis of PTSD was made according to Diagnostic and Statistical Manual of Mental Disorders (DSM–5) [53] criteria with the Clinician-Administered Posttraumatic Stress Disorder Scale for DSM-5 (CAPS-5) [54]. All controls in the PTSD cohort were trauma-exposed (based on DSM-5 criteria). Schizophrenia was diagnosed using the Structured Clinical Interview for DSM-IV (SCID) [55]. A diagnosis of PD was ascertained by a neurologist, according to UK Brain Bank Criteria [56].
Metabolic syndrome (MetS), which represents a cluster of risk factors for cardiovascular disease and type II diabetes, was assessed as an objective in the parent study. MetS has been found to significantly alter gene expression profiles [57] and was therefore considered as a covariate in the current study. Utilising the harmonised Joint Interim Statement (JIS) criteria [58], the presence of any three of the following five risk factors indicated a positive MetS diagnosis: (i) raised waist circumference (≥ 90 cm [59]); (ii) raised triglycerides (> 1.7 mmol/l); (iii) low HDL-C (men < 1.0 mmol/l, women < 1.3 mmol/l); (iv) raised blood pressure ≥ 130/85 mmHg or on hypertension treatment; and (v) raised fasting glucose ≥ 5.6 mmol/l or on diabetes treatment.
Childhood trauma was assessed using the Childhood Trauma Questionnaire (CTQ) [60]. Based on CTQ total score, the following cut-offs have been suggested: none to minimal trauma (25–36), low to moderate trauma (41–51), moderate to severe trauma (56–68) and severe to extreme trauma (73–125) [60]. We thus used a score ≥ 41 to identify individuals as having childhood trauma, as this represents the lowest level score indicative of abuse or neglect for each subscale. Major Depressive Disorder (MDD) was assessed using the MINI International Neuropsychiatric Interview, version 6.0 (MINI) [61]. Participants with current or lifetime MDD were classified as having MDD.
RNA isolation
Blood samples were collected from all participants using PAXgene Blood Collection tubes (Preanalytix, Switzerland), and RNA isolation was done using the PAXgene Blood RNA Kit (Qiagen, CA, USA). Samples were assessed for an RNA integrity number (RIN) using the Agilent 2100 Bioanalyzer (Agilent Technologies, California, USA). A RIN value of ≥ 7 is generally regarded as good quality, intact RNA [62].
RNA sequencing and analysis
Library preparation and sequencing
RNA-sequencing (RNA-seq) was performed at The Kinghorn Centre for Clinical Genomics (KCCG), Australia. Illumina’s TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold High Throughput was used for library preparation. Synthetic spike-ins (Sequins) were added at the point of the initial RNA dilution (1:50 dilution). Adapter dimers were detected in final library trace of the schizophrenia and PD samples, and all samples were therefore subjected to second bead clean-up, using a bead:sample ratio of 0.9. Following the bead clean-up, 14 samples (5 PD and 9 schizophrenia samples) still exhibited adapter dimers, and another bead clean-up procedure, using a bead:sample ratio of 0.85, was performed, after which the adapter dimers were removed. Since the differential treatment of the samples may introduce batch effects, we included the additional bead clean-up as a covariate in the investigations of the PD and schizophrenia cohorts.
Each sample was uniquely indexed, pooled and subsequently sequenced across ten lanes to minimise the potential for batch effects. Paired-end sequencing was performed on the Illumina HiSeq 2500 sequencing system, with a read length of 125 base pairs (bp) and sequencing depth of 50 million paired-end reads per sample.
RNA-seq mapping
The quality of the raw reads was assessed using FastQC tool (Version 0.1 1.5), and adaptor-end trimming was conducted using Trim Galore! (Version 0.4.2). Alignment of the RNA-seq reads to the Ensembl human reference genome (hg38/GRCH38) was performed using Spliced Transcripts Alignment to a Reference (STAR; Version 2.6.0c) [63]. The quantMode GeneCounts option was selected to generate raw gene-wise read counts for each sample.
Statistical analysis
The distributions of quantitative data were tested for normality using the Shapiro–Wilk test. Normal and asymmetrical data were summarised as mean and standard deviation, or median and interquartile range, respectively. Qualitative data were summarised as counts. To control for the effect of blood cell type composition variability on gene expression, blood cell type proportions were estimated using the ‘immunoStates’ expression matrix [64], and the DeconRNASeq package (317 genes across 20 cell types) [65] in R [66]. Cell types with the greatest composition variability have the potential to confound the gene expression results. Variables that exhibited significantly different distributions or frequencies between cases and controls in each diagnostic group were included as covariates in the gene expression analyses.
Exploratory analysis of the RNA-seq data was conducted using the R package DaMIR [67]. The DaMiR.sampleFilt function assesses the mean absolute correlation of each sample and removes those samples with a correlation lower than the value set in the th.corr argument. This was set at 0.9 in the current study.
We used the R package variancePartition [68] to assess the degree to which selected clinical and technical variables might influence gene expression and potentially confound the analysis. The package implements a linear mixed model method to characterise the contribution of selected variables to transcriptional variability. In the present study, total variance in gene expression was partitioned into the variance attributable to RIN, age, sex, nicotine use, and neutrophil and CD16 monocyte cell proportions in each cohort. Metabolic syndrome was included as a variable in the PTSD and PD cohorts, but not in the schizophrenia cohort (due to low numbers of MetS), and the additional bead clean-up was included as a variable in the PD and schizophrenia cohorts. Continuous variables (age, RIN, and neutrophil and CD16 monocyte proportions) were modelled as fixed effects, while categorical variables were modelled as random effects.
Differential expression analysis was performed using DESeq2 [69], which analyses differences in expression based on a negative binomial generalised linear model, adjusting for covariables as necessary. Statistical significance was assessed via the Wald test. The false discovery rate (FDR) was controlled for at 10% using the Benjamini–Hochberg method [70] for each pairwise comparison. Neutrophil and CD16 monocyte proportions, age, sex, RIN, and smoking status were included as covariates in all models. Metabolic syndrome was included as a covariate in the PTSD and PD models, but not the schizophrenia model, given the low frequency of MetS diagnosed in that cohort (12.5% and 5.6% in cases and controls, respectively (Table 1)). Current medication for hypertension was also included as a covariate in the PD cohort.
Table 1.
Clinical, demographic and haematological information for selected variables in the PTSD, schizophrenia and PD cohorts
| Demographic/clinical variable | PTSD (n = 79) | PDa (n = 30) | Schizophreniaa (n = 34) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | p | Case | Control | p | Case | Control | p | |
| (n = 40) | (n = 39) | (n = 12) | (n = 18) | (n = 16) | (n = 18) | ||||
| Age (years) (mean, SD) | 44.13 (9.38) | 45.32 (10.40) | 0.595 | 63.52 (7.29) | 61.93 (6.45) | 0.505 | 24.18 (4.79) | 23.4 (3.13) | 0.458 |
| Sex (Female, %) | 33 (82.5) | 31 (79.5) | 0.733 | 5 (41.7) | 7 (38.9) | 0.879 | 8 (50.0) | 8 (44.4) | 0.746 |
| Education (n, %) | |||||||||
| Grade 6 and below | 2 (5.0) | 0 (0) | 0.414d | 1 (8.3) | 0 (0) | 0.443d | 0 (0) | 0 (0) | 0.711d |
| Grades 7–9 | 12 (30.0) | 17 (43.6) | 5 (41.7) | 8 (44.4) | 5 (31.3) | 3 (16.7) | |||
| Grades 10–12 | 24 (60.0) | 20 (51.3) | 4 (33.3) | 9 (50.0) | 9 (56.3) | 11 (61.1) | |||
| Tertiary | 2 (5.0) | 2 (5.1) | 2 (16.7) | 1 (5.6) | 2 (12.5) | 4 (22.2) | |||
| Metabolic syndrome (yes, %) | 20 (50.0 | 19 (48.7) | 0.733 | 7 (58.3) | 10 (55.6) | 0.880 | 2 (12.5) | 1 (5.6) | 0.591d |
| Childhood trauma (yes, %) | 37 (92.5) | 24 (61.5) | 0.001b | 1 (8.3) | 4 (22.2) | 0.622d | 8 (50.0) | 12 (66.7) | 0.324 |
| CTQ score (median, IQR) | 51.5 (45.75–71.25) | 41 (32.5–56) | 0.002c | 32.5 (30.5–35.5) | 34.5 (30.25–39.75) | 0.337c | 40.5 (38.5–48.25) | 44.5 (40–57) | 0.276c |
| MDD (yes, %) | 24 (60.0) | 9 (23.1) | 0.001b | 0 (0) | 2 (11.1) | 0.503 | 3 (18.8) | 1 5.6) | 0.323d |
| Smoking (yes, %) | 25 (62.5) | 21 (53.8) | 0.436 | 5 (41.7) | 13 (72.2) | 0.094 | 12 (75.0) | 7 (38.9) | 0.034 |
| Alcohol (yes, %) | 32 (80.0) | 28 (71.8%) | 0.394 | 8 (66.7) | 15 (83.3) | 0.392d | 11 (68.8) | 11 (61.1) | 0.642 |
| Currently on anti-hypertensives (yes, %) | 10 (25.0) | 12 (30.8) | 0.567 | 8 (66.7) | 5 (27.8) | 0.061d | 0 (0%) | 0 (0) | 1 |
| CD16 Monocytee | 0.22 (0.08) | 0.23 (0.09) | 0.542 | 0.24 (0.11) | 0.28 (0.10) | 0.340 | 0.33 (0.07) | 0.29 (0.09) | 0.189 |
| Neutrophil (median, IQR) e | 0.36 (0.31–0.41) | 0.39 (0.32–0.42) | 0.202 | 0.348 (0.24–0.41) | 0.3065 (0.23–0.35) | 0.79c | 0.37 (0.26–0.42) | 0.32 (0.25–0.39) | 0.334 |
| RIN (medin, IQR) | 8.8 (8.4–9.0) | 8.8 (8.4–9.1) | 0.637c | 7.45 (6.95–8.1) | 7.6 (7.0–8.1) | 0.82c | 7.9 (7.7–8.2) | 7.8 (7.2–8.2) | 0.436c |
aExcluding outliers; bMann-Whitney U test; cKruskal-Wallis;dFisher's exact test; eestimated proportions; significant p-values (p < 0.05) indicated in bold
Weighted gene co-expression analysis was performed using the R package Co-Expression Modules identification Tool (CEMiTool) [71]. We applied the CEMiTool vst() function, which applies an arcsin-based variance stabilizing transformation if the correlation between the means and variances of genes is greater than 0.5 (vst) and also corrected for covariates included in the gene expression models for each NPD analysis using the empiricalBayesLM function implemented in the WGCNA package [72] prior to data input into CEMiTool.
As a first step, CEMiTool implements an unsupervised gene filtering step, based on the inverse gamma distribution. All default parameters were applied, with a variance filter p-value of 0.1. The beta-value (β), which represents the soft-thresholding power required to construct network adjacency, was selected using the default CEMiTool algorithm. Network construction was done using the unsigned method; in other words, up- and down regulated genes may be located in the same modules. We assume, therefore, that genes with a strong unsigned correlation are co-regulated. Gene-set enrichment analysis (GSEA) was performed using fgsea [73], implemented in CEMiTool, in order to identify the modules whose activity is associated with NPD status in each of the cohorts. Only the modules associated with each NPD were selected for further analysis.
Hub genes in each of the significantly associated modules were identified using the adjacency score in the CEMiTool R package. Module overrepresentation analysis (ORA) was performed based on the hypergeometric test, using the C2 Curated KEGG gene set list from the Molecular Signatures Database, version 7 (http://www.gsea-msigdb.org/gsea/msigdb/collections.jsp) [74]. Gene sets with an FDR < 0.05 were considered to be significantly overrepresented. To investigate molecular interactions between the genes within each of the NPD-associated modules, we superimposed module-wise co-expression networks onto human protein–protein interaction data downloaded from BioGRID (https://thebiogrid.org/), which makes use of peer-reviewed publications to manually curate gene and protein interaction data [75].
Results
Demographic and clinical information
Demographic and clinical variables for each diagnostic cohort are presented in Table 1. No significant differences in age between cases and controls in any of the diagnostic cohorts were observed. Individuals with PTSD were found to exhibit significantly more childhood trauma compared to trauma-exposed controls (TEC) (p = 0.001). Likewise, a significantly higher number of PTSD cases were found to have been diagnosed with MDD, compared to TECs (p = 0.001). Significantly more schizophrenia cases reported smoking cigarettes compared to controls (p = 0.034).
Disease-specific gene expression in PTSD, PD and schizophrenia cohorts
Four samples in the PD cohort (one male control, three female patients) and five samples in the schizophrenia cohort [three controls (one male, two female) and two male patients] were identified as outliers, and were therefore omitted from further analyses.
After filtering for genes with low expression (< 10 counts per gene) in each cohort, RNA-seq data for PTSD (n = 11,497 genes), PD (n = 11,763 genes) and SCZ (n = 10,710 genes), were assessed for the degree to which selected variables (e.g., RIN, age, sex, nicotine use, and neutrophil and CD16 monocytes) influenced gene expression and potentially confounded the NPD of interest (PTSD, PD or schizophrenia), using variancePartition [68].
The results indicated that, in the PTSD group, the proportion of neutrophils, CD16 monocytes and RIN accounted for the largest contribution to total gene expression variance, whereas MetS status, age, smoking and sex accounted for less of the gene expression variance and affected the expression of relatively fewer genes (Supplementary Figure 1(a) and (b)). Childhood trauma and MDD were not included as variables in the statistical model as they were both significantly associated with PTSD (Table 1) and may thus represent collinear variables.
Violin plots depicting the variance partition analysis in the PD cohort can be viewed in Supplementary Figure 2(a) and (b). Neutrophil and CD16 monocyte proportions, as well as RIN and age, were found to be the main drivers of gene expression variation, with current anti-hypertensive medication use, sex, smoking, metabolic syndrome status and second bead clean-up contributing to the variance in gene expression, although at a lower level.
As with the PD cohort, the top four drivers of gene expression variance in the schizophrenia cohort were found to be neutrophil and CD16 monocyte cell proportions, RIN and age, with sex, smoking, and the second bead clean-up contributing less (Supplementary Figure 3 (a) and (b)).
Only one gene, superoxide dismutase 2 (SOD2) (ENSG00000112096), was found to be differentially expressed between PTSD and TEC at a false discovery rate (FDR) < 0.1 (Supplementary Table 1). No genes were found to be differentially expressed at FDR < 0.1 in either the PD or the schizophrenia cohorts (Supplementary Tables 2 and 3, respectively).
Weighted gene co-expression network analysis (WGCNA)
Following default gene filtering by CEMiTool, 855, 833 and 911 genes were selected for analysis in the PTSD, PD and schizophrenia datasets, respectively. Using CEMiTool default settings, a soft-threshold β value of 14 (r2 = 0.87) was selected for PTSD, while a soft thresholds of 8 (r2 = 0.81) and 5 (r2 = 0.81) were selected for PD and schizophrenia, respectively.
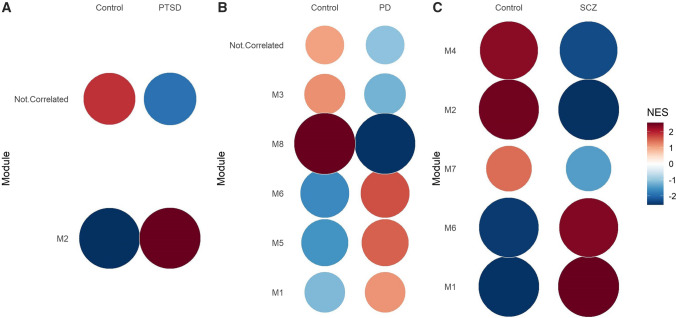
For the PTSD dataset, five modules were defined, ranging in size from 187 (Module 1) to 34 (Module 5) genes. Three hundred and twenty-five genes were defined as “not correlated”, i.e., they were not placed in any module (Supplementary Table 4). Module 2 (n = 158) was the only module found to be associated with PTSD (p = 3.26 × 10–16), following GSEA analysis (Fig. 1(a); Supplementary Table 4).
Fig. 1.
Gene set enrichment analysis showing the module activity for patient and control groups in the (A) PTSD, (B) PD and (C) schizophrenia (SCZ) cohorts. A Module 2 genes (n = 158) were found to have significantly increased expression in patients compared to controls in the PTSD cohort. B Genes in modules 1 (n = 222), 5 (n = 61), and 6 (n = 60) were found to be significantly increased in PD patients compared to controls, whereas those in modules 3 (n = 122) and 8 (n = 39) were significantly under-expressed in PD patients compared to controls. C Genes in modules 1 and 6 were found to be significantly increased in schizophrenia patients compared to schizophrenia controls, whereas those in modules 2, 4 and 7 were significantly under-expressed in schizophrenia patients compared to controls. M, module; SCZ, schizophrenia
Eight modules were identified in the PD cohort, ranging in size from 222 to 39 genes (Supplementary Table 5). Five of these modules were found to be significantly differentially expressed between PD patients and controls (Module 1 [n = 222; p < 0.001], Module 5 [n = 61; p < 0.001], Module 6 [n = 60; p < 0.001] and Module 8 [n = 39; p < 0.001] and Module 3 [n = 122; p = 0.001]) (Fig. 1 (b) and Supplementary Table 5).
We identified seven modules, ranging in size from 376 to 49 genes, in the schizophrenia cohort (Supplementary Table 6), and five of these were significantly associated with schizophrenia (Module 1 [n = 376, p = 6.45 × 10−45]; Module 2 [n = 185, p = 8.951 × 10−38]; Module 4 [n = 79, p = 5.80 × 10−21]; Module 6 [n = 63, p = 1.677 × 10−18] and Module 7 [n = 49, p = 8.006 × 10−5]) (Fig. 1(c) and Supplementary Table 6).
The normalised enrichment score (NES) represents the enrichment score for a module in each group, normalised by the number of genes in the module. The size and colour intensity of the circles correspond to the NES (Supplementary Tables 7–9).
Pathway enrichment analysis
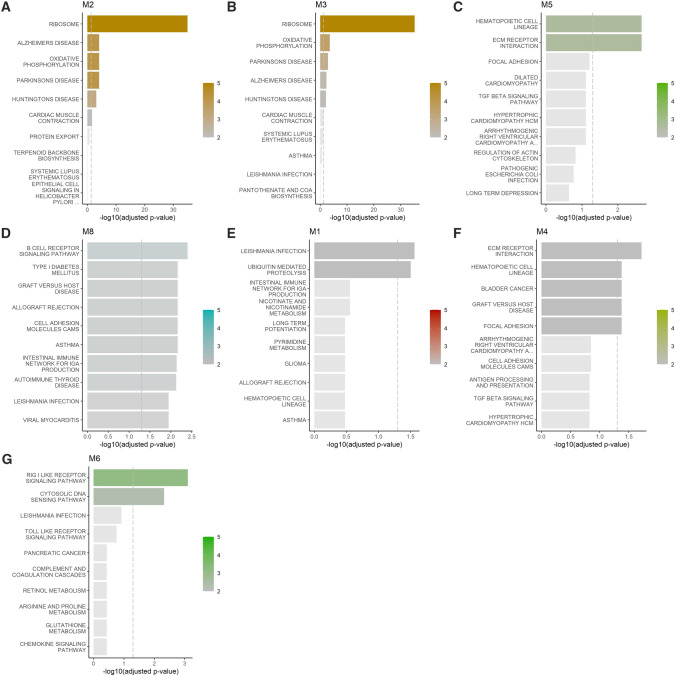
Overrepresentation analysis (ORA) of Module 2 genes in the PTSD cohort identified several enriched KEGG pathways, including Ribosome (p = 1.09 × 10–35), Parkinson’s Disease (p = 7.72 × 10−5), Oxidative phosphorylation (p = 7.72 × 10−5), Alzheimer’s Disease (p = 7.72 × 10−5), Huntington’s Disease (p = 8.40 × 10−3) and Cardiac muscle contraction pathways (p = 0.024) (Supplementary Table 10, Fig. 2(a)).
Fig. 2.
Over representation analysis for (A) PTSD, B–D PD and E–G schizophrenia. Bar graphs show the − log10 adjusted p-value of the enrichment between genes in modules and KEGG gene sets from MSigDB, the vertical dashed line indicates an adjusted p-value of 0.05. The colour intensities of the bars are proportional to the adjusted p-value
Overrepresented pathways were identified in three PD-associated gene modules (modules 3, 5 and 8) (Fig. 2B−D, Supplementary Table 11). The overrepresented KEGG pathways included Ribosome (p < 0.001), Oxidative phosphorylation (p < 0.001), Parkinson’s Disease (p = 0.001), Alzheimer’s Disease (p = 0.005) and Huntington’s Disease (p = 0.008) pathways in Module 3; ECM receptor interaction (p = 0.002), Hematopoietic cell lineage (p = 0.002) pathways in Module 5; and B-cell receptor signalling (p = 0.004), Asthma (p = 0.007), Cell adhesion molecules (p = 0.007), Allograft rejection (p = 0.007), Graft-versus-host disease (p = 0.007), Type I Diabetes mellitus (p = 0.007), Intestinal immune network for IgA production (p = 0.008), Autoimmune thyroid disease (p = 0.008), Viral myocarditis (p = 0.01), Leishmania infection (p = 0.012), Hematopoietic cell lineage (p = 0.014), Antigen processing and presentation (p = 0.014) and Systemic lupus erythematosus (p = 0.031) pathways (Module 8) (Supplementary Table 11).
Overrepresentation analysis of genes in schizophrenia-associated modules identified a number of enriched KEGG pathways in Module 1 (Leishmania infection (p = 0.030), Ubiquitin mediated proteolysis (p = 0.033)), Module 4 (ECM receptor interaction (p = 0.020), Focal adhesion (p = 0.044), Graft versus host disease (p = 0.044); Hematopoietic cell lineage (p = 0.044), Bladder cancer (p = 0.044)) and Module 6 (RIG I like receptor signalling pathway (p = 0.0008) and Cytosolic DNA sensing (p = 0.005)) (Fig. 2E−G, Supplementary Table 12). No KEGG pathways in modules 2 or 7 were found to be associated with schizophrenia.
Gene interaction analysis
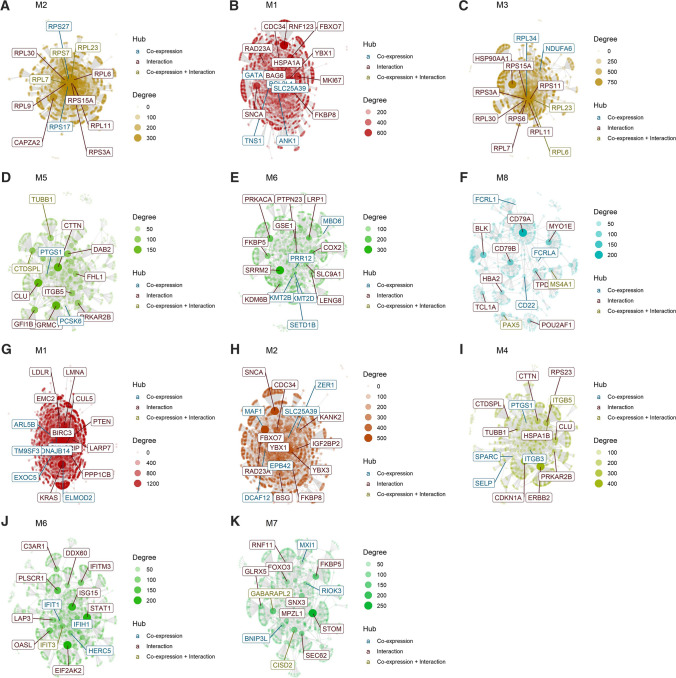
Hubs are defined as nodes (genes) with high connectivity (i.e., high number of neighbours) in the network. These represent nodes that could play an active role in the given context. Gene interaction analysis within PTSD-associated Module 2 identified 11 hub genes (RPS27, RPL6, RPL30, RPL11, RPL7, CAPZA2, RPL23, RPL9, RPS3A, RPS15, RPS7 and RPS17) (Fig. 3A). Likewise, a number of gene interactions were identified in the PD-associated modules (Fig. 3B–F) and schizophrenia-associated modules (Fig. 3G–K).
Fig. 3.
Interaction plots for each of the disorder-associated modules, indicating hub genes in each module. A Interaction plot for PTSD-associated Module 2, B–F Interaction plots for PD-associated modules 1, 3, 5, 6 and 8, and G–K Interaction plots for schizophrenia-associated modules 1, 2, 4, 6 and 7. The hubs are highlighted. The nodes represent the highly co-expressed genes within each module, in addition to those added by protein–protein interaction information, obtained from the BioGRID website. The genes are connected by co-expression and/or protein–protein interaction. Hubs are labelled and coloured based on their “origin”: if they are present in the CEMiTool module, they are coloured blue; if they have been inserted from the BioGRID interaction data, they are coloured red, and if they are shared between co-expressed and interaction databases, they are coloured yellow. The degree (connection strength) is reflected by the size of the node
Discussion
This is the first study to investigate the potential shared mechanisms underlying three common NPDs: PTSD, schizophrenia and PD. It is also, to the best of our knowledge, the first to present results from transcriptomic analyses on each of these NPDs in a Southern African population. Our main aim was to identify pathways underlying the disorders separately, and in combination, using a WGCNA approach.
We identified multiple common KEGG pathway signatures across PTSD Module 2 and PD Module 3, including the KEGG “Ribosome”, “Oxidative phosphorylation”, “Parkinson’s Disease”, “Huntington’s Disease” and “Alzheimer’s Disease” pathways. Ribosomes are subcellular organelles involved in the synthesis of proteins and comprise two subunits. The large subunit and smaller subunit (60S and 40S subunits, respectively, in eukaryotes) are encoded by ribosomal protein large (RPL) and ribosomal protein small (RPS) genes. The two subunits together form the 80S ribosomal nucleoprotein complex. Ribosomal proteins (RPs) are highly conserved and play an important role in the translation of proteins, specifically, it is thought, in the elongation step of protein synthesis, although the precise function is currently uncertain [76]. Translational homeostasis can be disturbed by alterations in translation initiation rates or RPs themselves, due to factors such as mutations or a decreased or increased number of normal ribosomes (altered gene expression or copy number). Disturbance in translational homeostasis can be pathologically significant in neurodegenerative disorders that are characterised by protein aggregation, one of the hallmark features of PD. Indeed, altered protein synthesis machinery has been found to be associated with alpha-synuclein oligomers in PD, although the expression of RPs was found to be stage- and region-dependent [77]. Interestingly, the downregulation of genes encoding RPs in peripheral tissue has recently been associated with aging [78], the main risk factor for PD [79], pointing towards the possibility of age-related translational dysfunction playing a role in susceptibility to PD. Although the literature on RPs and their association with PTSD is scarce, they have been found to play a role in other psychiatric disorders, including depression and obsessive–compulsive disorder (OCD) [80, 81]. However, the direction of this relationship, i.e., whether increased or decreased translational activity acts as a risk factor, may be disorder-specific.
Our observation that the ribosome pathway is dysregulated in blood in both PTSD and PD may point to dysfunctional protein homeostasis operating on a more systemic level. On the other hand, RPS3, a ribosomal protein that acts as a molecular switch for cell survival and death, has been found to be capable of traversing the blood–brain barrier, where it protected dopaminergic neurons in the substantia nigra from oxidative stress in a 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) mouse model. This was achieved by upregulating superoxide dismutase 1 (SOD1), which in turn scavenged the reactive oxygen species (ROS), preventing lipid peroxidation [82]. This indicates that alterations of RPs in the blood could affect CNS functioning.
It is important to note that, apart from their crucial role in protein synthesis, RPs also have numerous ribosome-independent functions, including immune signalling, tumorigenesis and development [83, 84]. To this end, RPs have been found to play a role in the interferon-gamma-mediated inflammatory response by modulating gene expression. For example, RPL13-A has been found to resolve inflammatory responses induced by IFN-gamma [83, 84] and RPS3 has been found to regulate genes that are involved in the NFKB immune response, as well as playing a role in cell survival and proliferation [85–87]. It is therefore plausible that altered ribosomal gene expression may result in dysregulation of immune pathways, and possibly contribute to the development of PTSD and/or PD in this way. Although the same pathways may be associated with both disorders, it is not possible to determine whether the same mechanisms within the pathway are disrupted in the disorders. It is, however, interesting that 50% of the PTSD Module 2 and PD Module 3 hub genes overlap, indicating that that some of the mechanisms may be similar.
Interestingly, RPL6, a hub gene in both PTSD Module 2 and PD Module 3, has been found to be downregulated in the peripheral blood mononucleocytes in PD patients [88, 89], and has been found to be associated with early-onset PD [90]. Anirudhan et al. [89] observed that RPL6 served as an intermediary, connecting three metalloprotein hubs associated with PD, and that the expression of RPL6 was regulated by serum levels of zinc and magnesium, which is notable given the evidence that metal exposure is an important risk factor for PD [91]. Although the data indicating an association between levels of metals and PTSD is currently sparse, zinc has been found to be associated with both depression and anxiety [92, 93], and serum levels of zinc have been found to be controlled through effective antidepressant pharmacotherapy [94]. In addition, an animal model of PTSD recently showed alterations in zinc concentration in selected brain regions of animals exhibiting a PTSD-like phenotype [95]. Magnesium has been found to play a role in depression, with an inverse relationship noted between dietary magnesium intake and depressive [96] and anxiety [97] symptoms, and the observation that lower serum levels of magnesium were correlated with increased depression as measured using the Patient Health Questionnaire [98]. Although the evidence for the role of magnesium in PTSD is, as with zinc, scarce, the aforementioned findings lay groundwork for further investigation into these relationships.
Genes comprising KEGG pathways of Focal adhesion and ECM receptor interaction were found to be enriched in the PD Module 5 but under-expressed in the schizophrenia Module 4. Both pathways have been implicated previously in schizophrenia [99–101] and PD [102, 103]. Focal adhesions are specialised regions in the cell where it interacts with the extracellular matrix (ECM). Interactions between cells and the ECM, mediated via transmembrane receptors such as integrins, facilitate the regulation of adhesion, migration and differentiation of cells. This is important in the maintenance of stable neuronal connectivity and the regulation of synaptic plasticity [104]. The involvement of the focal adhesion and ECM receptor interaction pathways in both PD and schizophrenia may point to the aberrant migration of neural progenitor cells during brain development in individuals who suffer from the schizophrenia, and dysregulation of adult synaptic plasticity and neuronal connectivity in PD.
We observed enrichment for several immune system related pathways in modules associated with PD, particularly in Module 8 (B-cell receptor signalling, Asthma, Allograft rejection, Graft vs host disease, Intestinal immune network for IgA production, Autoimmune thyroid disease, Antigen processing and presentation and Systemic lupus erythematosus KEGG pathways). Prior evidence indicates that chronic low-level inflammation, in both the nervous system and periphery, is a pathophysiological feature of PD [105], and increased cytokine levels observed in the clinical stages of disease suggest the involvement of peripheral inflammation in PD progression [106].
An altered gut microbiome has been found to be associated with PD [107, 108]. It is interesting to note that the ‘Intestinal immune network for IgA production’ pathway was found to be dysregulated in the PD cohort. The leucine-rich repeat kinase 2 (LRRK2) gene, which is one of the most frequent causes of familial PD [109, 110], is expressed in enteric neurons, where it has been found to modulate the IgA alterations via regulation of neuronal peptides [111]. IgA regulates commensal bacteria via a range of mechanisms, including enhancing bacterial-mucus interactions that aid in establishing and reinforcing the gut mucosal barrier, and suppressing bacterial species that stimulate the immune system and express genes that elicit an inflammatory response (reviewed in [112]). Thus, altered IgA-related gene expression in PD could lead to both altered microbial composition in favour of proinflammatory species, in turn potentially increasing peripheral inflammation, and could reduce the efficacy of the barrier to bacterial translocation. Disease-associated differences in IgA production could, therefore, provide a mechanism underlying recently published findings of microbiome differences in PD [113].
The KEGG ubiquitin mediated proteolysis pathway was found to be enriched in Module 1 of the schizophrenia cohort. The ubiquitin proteasome system (UPS) is a complex system that has important roles in diverse processes, including protein labelling for degradation, vesicle and protein transport, membrane receptor recycling, and modulation of cellular responses to inflammation and oxidative stress [114, 115]. Importantly, the UPS has also been described as a crucial regulator of neural development and the maintenance of brain structure and function [116], and has been found to be associated with schizophrenia at a transcriptomic and proteomic level, in both peripheral and central tissue [117–124]. In addition, ubiquitin-conjugating enzyme (UBE2K) (a hub gene of schizophrenia Module 1 ubiquitin-mediated proteolysis pathway) has been found to be associated with positive symptom domains of psychosis [121]. Dysregulation of the proteasome system could result in accumulation of dysfunctional and/or damaged proteins. To this end, recent evidence showed that accumulation of ubiquitinated proteins has been detected in a subgroup of schizophrenia patients [125, 126], and it is thought that this may characterise a biological subtype of schizophrenia [124]. This finding represents an interesting point of departure for the current study and should be further investigated in a larger schizophrenia cohort.
No differentially expressed genes were identified in either the PD or the schizophrenia cohorts, and only one gene (superoxide dismutase 2, SOD2) was identified as being differentially expressed between patients and TECs in the PTSD cohort. SOD2, also known as manganese-dependent SOD (MnSOD), is an enzyme that plays an important role as a first line of defence against ROS, and in doing so, protects mitochondria from oxidative damage. ROS perform numerous vital functions in the cell, and especially in the CNS, where they play an important role in normal brain physiology [127]. In healthy individuals, the production of ROS is tightly regulated by, amongst others, an enzymatic defence mechanism, facilitated by antioxidant enzymes such as the SOD family of enzymes, catalase and glutathione peroxidase [128]. Oxidative stress (OXS) is a result of an imbalance in ROS and antioxidant mechanisms responsible for “mopping” the ROS up, and results in detrimental effects, including damage to proteins, lipids and DNA, aberrant downstream signalling, and the stimulation of apoptosis [129–132]. OXS has been associated with neurodegenerative disorders, accelerated cellular aging, vascular disorders, cancers and numerous neuropsychiatric disorders [127, 133]. Recent evidence points towards the association between OXS and PTSD, with animal and human studies implicating increased levels of ROS and OXS in the development of the disorder [127, 134, 135]. The SOD enzymes have been found to be associated with PTSD, although not all results have been consistent, which may reflect differences in diagnostic criteria and research aims. Tezcan et al. [136] observed a positive correlation between total SOD levels in hemolysates and PTSD CAPS scores. On the other hand, Borovac Štefanović et al. [137] observed reduced SOD levels in PTSD compared to healthy controls, and Zieker et al. [138] observed reduced expression of SOD1 in PTSD compared to control individuals. Although the results from the present study require replication in a larger sample, the increased SOD2 expression observed in the present study may reflect increased cytokine production in PTSD patients, and an attempt to protect the cells from OXS, since increased cytokine production has been found to result in the induction of nitric oxide and ROS [139, 140].
The current results need to be viewed in the context of some important limitations. The use of blood instead of brain tissue, although unavoidable in our setting, is a limitation. Blood tissue is complex, comprising a number of subsets of cell populations, each with their own gene expression profile. However, using cell type deconvolution methods, we did not observe significant differences in cell type proportions between cases and controls within each diagnostic cohort. In addition, blood and brain gene expression profiles have been found to correlate, although somewhat variably, in different studies [141–146]. PTSD, schizophrenia, and PD have also been found to possess underlying immunological disturbances, which highlights the value and practicality of using blood samples in gene expression studies for these disorders. Two of the three disorders investigated, PD and schizophrenia, had very small sample sizes, resulting in a lack of statistical power, which may have prevented identification of more subtle gene expression signatures. Although the pathway-based approach provides a broader overview of biological mechanisms, the pathways implicated are intricate, complex and composed of multiple different genes. Further mechanistic studies are required to identify the key components in these pathways, their individual and interactive effects on the molecular level, and their influence on disorder phenotype. Integration of the current RNAseq data with additional -omics investigations, such as DNA methylation and proteomic investigations would have shed light on potential key players in the associated pathways. DNA methylation data are in the process of being analyzed for the PTSD cohort and will be included in a future manuscript.
This study also has several important strengths. Transcriptomic studies provide a measure of functional data that is not afforded by other genetic investigations, such as GWAS. By identifying shared and distinct gene expression profiles associated with these three disorders, our study is aligned with the movement seeking to identify transdiagnostic mechanisms that can bridge the previously siloed investigations of NPDs. Indeed, our results not only point to the complex and multicausal nature of NPDs, but also offer insight into key mechanisms that could be targeted to produce pleiotropic effects on neurobiology and behaviour. This study was also conducted in an African ancestry population. This is relevant given calls to increase the diversity of representation in genetic studies in general, and psychiatric genetic studies, in particular. Furthermore, in keeping with the gene-environment framework, local contexts can influence the risk and course of disorders. For this reason, the results of locally conducted studies are particularly relevant.
In conclusion, we adopted a pathway-based approach to identify overarching mechanisms shared by PTSD, schizophrenia and PD. Our analyses implicated widely acting biological processes influencing ribosome machinery, inflammation, and ubiquitination, all of which have broad scope to affect behaviour. While our data provide preliminary evidence for shared molecular aetiology between PTSD and PD, and between PD and schizophrenia, the results require replication in a larger sample.
Supplementary Information
Below is the link to the electronic supplementary material.
Authors' contributions
Conceptualization: SMJH, SS; methodology: SMJH, PS, SM, GT; formal analysis and investigation: SMJH, PS; writing—original draft preparation: SMJH, PS, JSW, SA, NWM; writing—manuscript editing: SMJH, PS, JSW, ESO, LLH, NM, SM, SB, SA, GT, RE, JC, SS. All authors read and approved the final manuscript.
Funding
Research reported in this publication was supported by the South African Medical Research Council for the “Shared Roots” Flagship Project, Grant no. MRC-RFA-IFSP-01-2013/SHARED ROOTS” through funding received from the South African National Treasury under its Economic Competitiveness and Support Package. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the South African Medical Research Council. The work by Leigh van den Heuvel reported herein was made possible through funding by the South African Medical Research Council through its Division of Research Capacity Development under the SAMRC Clinician Researcher (M.D PhD) Scholarship Programme from funding received from the South African National Treasury. The content hereof is the sole responsibility of the authors and do not necessarily represent the official views of the SAMRC or the funders.
Data availability
The datasets generated during and/or analysed during the current study are not publicly available due to lack of participant informed consent and ethics approval to deposit the data in a public repository, but are available from the corresponding author on reasonable request.
Code availability
The code generated during the current study is available from the corresponding author on request.
Declarations
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Competing interests
Soraya Seedat is a member of the Editorial Board of Discover Mental Health. She was not involved in the peer-review or handling of the manuscript. The authors have no other competing interests to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vos T, Flaxman AD, Naghavi M, Lozano R, Michaud C, Ezzati M, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2163–2196. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vos T, Lim SS, Abbafati C, Abbas KM, Abbasi M, Abbasifard M, et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396:1204–1222. doi: 10.1016/S0140-6736(20)30925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breslau N. The epidemiology of trauma, PTSD, and other posttrauma disorders. Trauma Violence Abuse. 2009;10:198–210. doi: 10.1177/1524838009334448. [DOI] [PubMed] [Google Scholar]
- 4.Williams SL, Williams DR, Stein DJ, Seedat S, Jackson PB, Moomal H. Multiple traumatic events and psychological distress: the South Africa stress and health study. J Trauma Stress. 2007;20:845–855. doi: 10.1002/jts.20252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charlson FJ, Ferrari AJ, Santomauro DF, Diminic S, Stockings E, Scott JG, et al. Global epidemiology and burden of schizophrenia: findings from the Global Burden of Disease Study 2016. Schizophr Bull. 2018;44:1195–1203. doi: 10.1093/schbul/sby058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mueser KT, McGurk SR. Schizophrenia Lancet. 2004;363:2063–2072. doi: 10.1016/S0140-6736(04)16458-1. [DOI] [PubMed] [Google Scholar]
- 7.Tandon R, Nasrallah HA, Keshavan MS. Schizophrenia, “just the facts” 4. Clinical features and conceptualization. Schizophr Res. 2009;110:1–23. doi: 10.1016/j.schres.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Laursen TM, Nordentoft M, Mortensen PB. Excess early mortality in schizophrenia. Annu Rev Clin Psychol. 2014;10:425–448. doi: 10.1146/annurev-clinpsy-032813-153657. [DOI] [PubMed] [Google Scholar]
- 9.GBD 2015 Disease and Injury Incidence and Prevalence Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–602. doi: 10.1016/S0140-6736(16)31678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng H, Wang P, Jankovic J. The genetics of Parkinson disease. Ageing Res Rev. 2018;42:72–85. doi: 10.1016/j.arr.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S, Ressler KJ. Genomic updates in understanding PTSD. Prog Neuropsychopharmacol Biol Psychiatry. 2019;90:197–203. doi: 10.1016/j.pnpbp.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golov AK, Kondratyev NV, Kostyuk GP, Golimbet VE. Novel approaches for identifying the molecular background of schizophrenia. Cells. 2020;9:246. doi: 10.3390/cells9010246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kouli A, Torsney KM, Kuan W-L. Parkinson’s disease: etiology, neuropathology, and pathogenesis. In: Stoker TB, Greenland JC, editors. Parkinson’s disease: pathogenesis and clinical aspects. Brisbane (AU): Codon Publications; 2018. [PubMed] [Google Scholar]
- 14.Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen C-Y, Choi KW, et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun. 2019;10:4558. doi: 10.1038/s41467-019-12576-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swart PC, van den Heuvel LL, Lewis CM, Seedat S, Hemmings SMJ. A genome-wide association study and polygenic risk score analysis of posttraumatic stress disorder and metabolic syndrome in a south african population. Front Neurosci. 2021;15:677800. doi: 10.3389/fnins.2021.677800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg MS, Tanev K, Marin M-F, Pitman RK. Stress, PTSD, and dementia. Alzheimer’s Dementia. 2014;10:S155–S165. doi: 10.1016/j.jalz.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Khachaturian AS, Khachaturian ZS. Military risk factors for Alzheimer’s dementia and neurodegenerative disease. Alzheimer’s Dementia. 2014;10:S90–S91. doi: 10.1016/j.jalz.2014.05.1085. [DOI] [PubMed] [Google Scholar]
- 18.Chan Y-LE, Bai Y-M, Hsu J-W, Huang K-L, Su T-P, Li C-T, et al. Post-traumatic stress disorder and risk of parkinson disease: a nationwide longitudinal study. Am J Geriatr Psychiatry. 2017;25:917–23. doi: 10.1016/j.jagp.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Svensson E, Farkas DK, Gradus JL, Lash TL, Sørensen HT. Adjustment disorder and risk of Parkinson’s disease. Eur J Neurol. 2016;23:751–756. doi: 10.1111/ene.12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucassen PJ, Pruessner J, Sousa N, Almeida OFX, Van Dam AM, Rajkowska G, et al. Neuropathology of stress. Acta Neuropathol. 2014;127:109–135. doi: 10.1007/s00401-013-1223-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torrey EF. Studies of individuals with schizophrenia never treated with antipsychotic medications: a review. Schizophr Res. 2002;58:101–115. doi: 10.1016/s0920-9964(02)00381-x. [DOI] [PubMed] [Google Scholar]
- 22.Pappa S, Dazzan P. Spontaneous movement disorders in antipsychotic-naive patients with first-episode psychoses: a systematic review. Psychol Med. 2009;39:1065–1076. doi: 10.1017/S0033291708004716. [DOI] [PubMed] [Google Scholar]
- 23.Waddington JL. Psychosis in Parkinson’s disease and parkinsonism in antipsychotic-naive schizophrenia spectrum psychosis: clinical, nosological and pathobiological challenges. Acta Pharmacologica Sinica. 2020 doi: 10.1038/s41401-020-0373-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf RC, Rashidi M, Fritze S, Kubera KM, Northoff G, Sambataro F, et al. A neural signature of parkinsonism in patients with schizophrenia spectrum disorders: a multimodal MRI study using parallel ICA. Schizophr Bull. 2020;46:999–1008. doi: 10.1093/schbul/sbaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forsaa EB, Larsen JP, Wentzel-Larsen T, Goetz CG, Stebbins GT, Aarsland D, et al. A 12-year population-based study of psychosis in Parkinson disease. Arch Neurol. 2010;67:996–1001. doi: 10.1001/archneurol.2010.166. [DOI] [PubMed] [Google Scholar]
- 26.Pagonabarraga J, Martinez-Horta S, de Bobadilla FR, Pérez J, Ribosa-Nogué R, Marín J, et al. Minor hallucinations occur in drug-naive Parkinson’s disease patients, even from the premotor phase. Mov Disord. 2016;31:45–52. doi: 10.1002/mds.26432. [DOI] [PubMed] [Google Scholar]
- 27.Schapira AHV, Chaudhuri KR, Jenner P. Non-motor features of Parkinson disease. Nat Rev Neurosci. 2017;18:435–450. doi: 10.1038/nrn.2017.62. [DOI] [PubMed] [Google Scholar]
- 28.Lenka A, Pagonabarraga J, Pal PK, Bejr-Kasem H, Kulisvesky J. Minor hallucinations in Parkinson disease: a subtle symptom with major clinical implications. Neurology. 2019;93:259–266. doi: 10.1212/WNL.0000000000007913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smeland OB, Shadrin A, Bahrami S, Broce I, Tesli M, Frei O, et al. Genome-wide association analysis of Parkinson’s disease and schizophrenia reveals shared genetic architecture and identifies novel risk loci. Biol Psychiatry. 2020 doi: 10.1016/j.biopsych.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48:709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, van der Brug M, Cai F, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassett AS, Chow EWC, AbdelMalik P, Gheorghiu M, Husted J, Weksberg R. The schizophrenia phenotype in 22q11 deletion syndrome. Am J Psychiatry. 2003;160:1580–1586. doi: 10.1176/appi.ajp.160.9.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaleski C, Bassett AS, Tam K, Shugar AL, Chow EWC, McPherson E. The co-occurrence of early onset Parkinson disease and 22q11.2 deletion syndrome. Am J Med Genet A. 2009;149A:525–8. doi: 10.1002/ajmg.a.32650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Booij J, van Amelsvoort T, Boot E. Co-occurrence of early-onset Parkinson disease and 22q11.2 deletion syndrome: potential role for dopamine transporter imaging. Am J Med Genet A. 2010;152A:2937–8. doi: 10.1002/ajmg.a.33665. [DOI] [PubMed] [Google Scholar]
- 38.Butcher NJ, Kiehl T-R, Hazrati L-N, Chow EWC, Rogaeva E, Lang AE, et al. Association between early-onset Parkinson disease and 22q11.2 deletion syndrome: identification of a novel genetic form of Parkinson disease and its clinical implications. JAMA Neurol. 2013;70:1359–66. doi: 10.1001/jamaneurol.2013.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehman AF, Dhamija R, Williams ES, Barrett MJ. 22q11.2 deletion syndrome presenting with early-onset Parkinson’s disease. Mov Disord. 2015;30:1289–90. doi: 10.1002/mds.26305. [DOI] [PubMed] [Google Scholar]
- 40.Weinstein JJ, Chohan MO, Slifstein M, Kegeles LS, Moore H, Abi-Dargham A. Pathway-specific dopamine abnormalities in schizophrenia. Biol Psychiatry. 2017;81:31–42. doi: 10.1016/j.biopsych.2016.03.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masato A, Plotegher N, Boassa D, Bubacco L. Impaired dopamine metabolism in Parkinson’s disease pathogenesis. Mol Neurodegener. 2019;14:35. doi: 10.1186/s13024-019-0332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kessler RC. Posttraumatic stress disorder in the national comorbidity survey. Arch Gen Psychiatry. 1995;52:1048. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- 43.Buckley PF, Miller BJ, Lehrer DS, Castle DJ. Psychiatric comorbidities and schizophrenia. Schizophr Bull. 2009;35:383–402. doi: 10.1093/schbul/sbn135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Achim AM, Maziade M, Raymond É, Olivier D, Mérette C, Roy M-A. How prevalent are anxiety disorders in schizophrenia? A meta-analysis and critical review on a significant association. Schizophr Bull. 2011;37:811–821. doi: 10.1093/schbul/sbp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamner MB, Frueh BC, Ulmer HG, Huber MG, Twomey TJ, Tyson C, et al. Psychotic features in chronic posttraumatic stress disorder and schizophrenia: comparative severity. J Nerv Ment Dis. 2000;188:217–221. doi: 10.1097/00005053-200004000-00004. [DOI] [PubMed] [Google Scholar]
- 46.Sareen J, Cox BJ, Goodwin RD, Asmundson GJG. Co-occurrence of posttraumatic stress disorder with positive psychotic symptoms in a nationally representative sample. J Trauma Stress. 2005;18:313–22. doi: 10.1002/jts.20040. [DOI] [PubMed] [Google Scholar]
- 47.Braakman MH, Kortmann FAM, van den Brink W. Validity of “post-traumatic stress disorder with secondary psychotic features”: a review of the evidence. Acta Psychiatr Scand. 2009;119:15–24. doi: 10.1111/j.1600-0447.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- 48.Blevins CA, Weathers FW, Witte TK. Dissociation and posttraumatic stress disorder: a latent profile analysis. J Trauma Stress. 2014;27:388–396. doi: 10.1002/jts.21933. [DOI] [PubMed] [Google Scholar]
- 49.Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE, et al. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol Psychiatry. 2018;23:666–673. doi: 10.1038/mp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. PNAS. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang D, Tang C, Zhang A. Cluster analysis for gene expression data: a survey. IEEE Trans Knowl Data Eng. 2004;16:1370–1386. doi: 10.1109/TKDE.2004.68. [DOI] [Google Scholar]
- 52.Pierson E, Consortium the Gte. Koller D, Battle A, Mostafavi S. Sharing and specificity of co-expression networks across 35 human tissues. PLOS Comput Biol. 2015;11:e1004220. doi: 10.1371/journal.pcbi.1004220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-5. 5th edition. Arlington, VA: American Psychiatric Association; n.d.
- 54.Weathers F, Blake D, Schnurr P, Kaloupek D, Marx B, Keane T. The clinician-administered PTSD scale for DSM-5 (CAPS-5) 2013. [DOI] [PMC free article] [PubMed]
- 55.First MB, Spitzer RL, Gobbon M, Williams JBW. Structured clinical interview for DSM-IV Axis I disorders–Patient Edition (SCID-I/P, Version 2.0, 8/98 revision). New York: New York State Psychiatric Institute, Biometrics Department; 1998.
- 56.Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55:181–184. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grayson BL, Wang L, Aune TM. Peripheral blood gene expression profiles in metabolic syndrome, coronary artery disease and type 2 diabetes. Genes Immun. 2011;12:341–351. doi: 10.1038/gene.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alberti KGMM, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 59.Matsha TE, Hassan MS, Hon GM, Soita DJ, Kengne AP, Erasmus RT. Derivation and validation of a waist circumference optimal cutoff for diagnosing metabolic syndrome in a South African mixed ancestry population. Int J Cardiol. 2013;168:2954–2955. doi: 10.1016/j.ijcard.2013.03.150. [DOI] [PubMed] [Google Scholar]
- 60.Bernstein DP, Fink L. Childhood trauma questionnaire: a retrospective self-report manual. San Antonio: Psychological Corporation; 1998. [Google Scholar]
- 61.Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry. 1998;59(Suppl 2):22–33. [PubMed] [Google Scholar]
- 62.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vallania F, Tam A, Lofgren S, Schaffert S, Azad TD, Bongen E, et al. Leveraging heterogeneity across multiple datasets increases cell-mixture deconvolution accuracy and reduces biological and technical biases. Nat Commun. 2018;9:4735. doi: 10.1038/s41467-018-07242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gong T, Szustakowski JD. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics. 2013;29:1083–1085. doi: 10.1093/bioinformatics/btt090. [DOI] [PubMed] [Google Scholar]
- 66.R Core Team. R : A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2020.
- 67.Chiesa M, Colombo GI, Piacentini L. DaMiRseq—an R/Bioconductor package for data mining of RNA-Seq data: normalization, feature selection and classification. Bioinformatics. 2018;34:1416–1418. doi: 10.1093/bioinformatics/btx795. [DOI] [PubMed] [Google Scholar]
- 68.Hoffman GE, Schadt EE. variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinform. 2016;17:483. doi: 10.1186/s12859-016-1323-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benjamini Y, Hochberg Y. Controlling the false-discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- 71.Russo PST, Ferreira GR, Cardozo LE, Bürger MC, Arias-Carrasco R, Maruyama SR, et al. CEMiTool: a bioconductor package for performing comprehensive modular co-expression analyses. BMC Bioinform. 2018;19:56. doi: 10.1186/s12859-018-2053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Korotkevich G, Sukhov V, Sergushichev A. Fast gene set enrichment analysis. BioRxiv. 2019 doi: 10.1101/060012. [DOI] [Google Scholar]
- 74.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oughtred R, Rust J, Chang C, Breitkreutz B-J, Stark C, Willems A, et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grela P, Sawa-Makarska J, Gordiyenko Y, Robinson CV, Grankowski N, Tchórzewski M. Structural properties of the human acidic ribosomal P proteins forming the P1–P2 heterocomplex. J Biochem. 2008;143:169–177. doi: 10.1093/jb/mvm207. [DOI] [PubMed] [Google Scholar]
- 77.Garcia-Esparcia P, Hernández-Ortega K, Koneti A, Gil L, Delgado-Morales R, Castaño E, et al. Altered machinery of protein synthesis is region- and stage-dependent and is associated with α-synuclein oligomers in Parkinson’s disease. Acta Neuropathol Commun. 2015;3:76. doi: 10.1186/s40478-015-0257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, et al. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. doi: 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reeve A, Simcox E, Turnbull D. Ageing and Parkinson’s disease: why is advancing age the biggest risk factor? Ageing Res Rev. 2014;14:19–30. doi: 10.1016/j.arr.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smagin DA, Kovalenko IL, Galyamina AG, Bragin AO, Orlov YL, Kudryavtseva NN. Dysfunction in ribosomal gene expression in the hypothalamus and hippocampus following chronic social defeat stress in male mice as revealed by RNA-Seq. Neural Plast. 2016;2016:e3289187. doi: 10.1155/2016/3289187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song Y, Liu Y, Wu P, Zhang F, Wang G. Genome-wide mRNA expression analysis of peripheral blood from patients with obsessive-compulsive disorder. Sci Rep. 2018 doi: 10.1038/s41598-018-30624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ahn EH, Kim DW, Shin MJ, Kim YN, Kim HR, Woo SJ, et al. PEP-1-ribosomal protein S3 protects dopaminergic neurons in an MPTP-induced Parkinson’s disease mouse model. Free Radic Biol Med. 2013;55:36–45. doi: 10.1016/j.freeradbiomed.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 83.Zhou X, Liao W-J, Liao J-M, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poddar D, Basu A, Baldwin WM, Kondratov RV, Barik S, Mazumder B. An Extraribosomal function of ribosomal protein L13a in macrophages resolves inflammation. J Immunol. 2013;190:3600–3612. doi: 10.4049/jimmunol.1201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, et al. Ribosomal protein S3: a KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 86.Lee SB, Kwon I-S, Park J, Lee K-H, Ahn Y, Lee C, et al. Ribosomal protein S3, a new substrate of Akt, serves as a signal mediator between neuronal apoptosis and DNA repair. J Biol Chem. 2010;285:29457–29468. doi: 10.1074/jbc.M110.131367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wan F, Lenardo MJ. The nuclear signaling of NF-κB: current knowledge, new insights, and future perspectives. Cell Res. 2010;20:24–33. doi: 10.1038/cr.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duarte RRR, Bachtel ND, Côtel M-C, Lee SH, Selvackadunco S, Watson IA, et al. The psychiatric risk gene NT5C2 regulates adenosine monophosphate-activated protein kinase signaling and protein translation in human neural progenitor cells. Biol Psychiatry. 2019;86:120–130. doi: 10.1016/j.biopsych.2019.03.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Anirudhan A, Angulo-Bejarano PI, Paramasivam P, Manokaran K, Kamath SM, Murugesan R, et al. RPL6: a key molecule regulating zinc- and magnesium-bound metalloproteins of Parkinson’s disease. Front Neurosci. 2021 doi: 10.3389/fnins.2021.631892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hamed M, Gladbach Y, Möller S, Fischer S, Ernst M, Struckmann S, et al. A workflow for the integrative transcriptomic description of molecular pathology and the suggestion of normalizing compounds, exemplified by Parkinson’s disease. Sci Rep. 2018;8:7937. doi: 10.1038/s41598-018-25754-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goldman SM. Environmental toxins and Parkinson’s disease. Annu Rev Pharmacol Toxicol. 2014;54:141–164. doi: 10.1146/annurev-pharmtox-011613-135937. [DOI] [PubMed] [Google Scholar]
- 92.Whittle N, Lubec G, Singewald N. Zinc deficiency induces enhanced depression-like behaviour and altered limbic activation reversed by antidepressant treatment in mice. Amino Acids. 2009;36:147–158. doi: 10.1007/s00726-008-0195-6. [DOI] [PubMed] [Google Scholar]
- 93.Młyniec K, Davies CL, Budziszewska B, Opoka W, Reczyński W, Sowa-Kućma M, et al. Time course of zinc deprivation-induced alterations of mice behavior in the forced swim test. Pharmacol Rep. 2012;64:567–575. doi: 10.1016/s1734-1140(12)70852-6. [DOI] [PubMed] [Google Scholar]
- 94.Takeda A, Tamano H, Kan F, Itoh H, Oku N. Anxiety-like behavior of young rats after 2-week zinc deprivation. Behav Brain Res. 2007;177:1–6. doi: 10.1016/j.bbr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 95.Sela H, Cohen H, Karpas Z, Zeiri Y. Distinctive hippocampal zinc distribution patterns following stress exposure in an animal model of PTSD. Metallomics. 2017;9:323–333. doi: 10.1039/c6mt00207b. [DOI] [PubMed] [Google Scholar]
- 96.Sun C, Wang R, Li Z, Zhang D. Dietary magnesium intake and risk of depression. J Affect Disord. 2019;246:627–632. doi: 10.1016/j.jad.2018.12.114. [DOI] [PubMed] [Google Scholar]
- 97.Anjom-Shoae J, Sadeghi O, Hassanzadeh Keshteli A, Afshar H, Esmaillzadeh A, Adibi P. The association between dietary intake of magnesium and psychiatric disorders among Iranian adults: a cross-sectional study. Br J Nutr. 2018;120:693–702. doi: 10.1017/S0007114518001782. [DOI] [PubMed] [Google Scholar]
- 98.Tarleton EK, Kennedy AG, Rose GL, Crocker A, Littenberg B. The association between serum magnesium levels and depression in an adult primary care population. Nutrients. 2019;11:1475. doi: 10.3390/nu11071475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berretta S. Extracellular matrix abnormalities in schizophrenia. Neuropharmacology. 2012;62:1584–1597. doi: 10.1016/j.neuropharm.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fan Y, Abrahamsen G, Mills R, Calderón CC, Tee JY, Leyton L, et al. Focal adhesion dynamics are altered in schizophrenia. Biol Psychiatry. 2013;74:418–426. doi: 10.1016/j.biopsych.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 101.Tee JY, Sutharsan R, Fan Y, Mackay-Sim A. Cell migration in schizophrenia: patient-derived cells do not regulate motility in response to extracellular matrix. Mol Cell Neurosci. 2017;80:111–122. doi: 10.1016/j.mcn.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 102.Edwards YJK, Beecham GW, Scott WK, Khuri S, Bademci G, Tekin D, et al. Identifying consensus disease pathways in Parkinson’s disease using an integrative systems biology approach. PLoS ONE. 2011;6:e16917. doi: 10.1371/journal.pone.0016917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shen J, Chen X-C, Li W-J, Han Q, Chen C, Lu J-M, et al. Identification of Parkinson’s disease-related pathways and potential risk factors. J Int Med Res. 2020;48:300060520957197. doi: 10.1177/0300060520957197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McGlashan TH, Hoffman RE. Schizophrenia as a disorder of developmentally reduced synaptic connectivity. Arch Gen Psychiatry. 2000;57:637–648. doi: 10.1001/pubs.ArchGenPsychiatry-ISSN-0003-990x-57-7-ynv9397. [DOI] [PubMed] [Google Scholar]
- 105.Wallings RL, Herrick MK, Tansey MG. LRRK2 at the interface between peripheral and central immune function in Parkinson’s. Front Neurosci. 2020;14:443. doi: 10.3389/fnins.2020.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reale M, Iarlori C, Thomas A, Gambi D, Perfetti B, Di Nicola M, et al. Peripheral cytokines profile in Parkinson’s disease. Brain Behav Immun. 2009;23:55–63. doi: 10.1016/j.bbi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 107.Aho VTE, Pereira PAB, Voutilainen S, Paulin L, Pekkonen E, Auvinen P, et al. Gut microbiota in Parkinson’s disease: Temporal stability and relations to disease progression. EBioMedicine. 2019;44:691–707. doi: 10.1016/j.ebiom.2019.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin C-H, Chen C-C, Chiang H-L, Liou J-M, Chang C-M, Lu T-P, et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J Neuroinflamm. 2019;16:129. doi: 10.1186/s12974-019-1528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Healy DG, Falchi M, O’Sullivan SS, Bonifati V, Durr A, Bressman S, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: a case-control study. Lancet Neurol. 2008;7:583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lunati A, Lesage S, Brice A. The genetic landscape of Parkinson’s disease. Rev Neurol (Paris) 2018;174:628–643. doi: 10.1016/j.neurol.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 111.Maekawa T, Shimayama H, Tsushima H, Kawakami F, Kawashima R, Kubo M, et al. LRRK2: an emerging new molecule in the enteric neuronal system that quantitatively regulates neuronal peptides and iga in the gut. Dig Dis Sci. 2017;62:903–912. doi: 10.1007/s10620-017-4476-3. [DOI] [PubMed] [Google Scholar]
- 112.Suzuki K. Diversified IgA–bacteria interaction in gut homeostasis. In: Wang J-Y, editor. B cells in immunity and tolerance, vol. 1254, Singapore: Springer Singapore; 2020, p. 105–16. 10.1007/978-981-15-3532-1_9. [DOI] [PubMed]
- 113.Romano S, Savva GM, Bedarf JR, Charles IG, Hildebrand F, Narbad A. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. Npj Parkinsons Dis. 2021;7:1–13. doi: 10.1038/s41531-021-00156-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 115.Haas KF, Broadie K. Roles of ubiquitination at the synapse. Biochim Biophys Acta. 2008;1779:495–506. doi: 10.1016/j.bbagrm.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Luza S, Opazo CM, Bousman CA, Pantelis C, Bush AI, Everall IP. The ubiquitin proteasome system and schizophrenia. Lancet Psychiatry. 2020;7:528–537. doi: 10.1016/S2215-0366(19)30520-6. [DOI] [PubMed] [Google Scholar]
- 117.Vawter MP, Barrett T, Cheadle C, Sokolov BP, Wood WH, Donovan DM, et al. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res Bull. 2001;55:641–650. doi: 10.1016/s0361-9230(01)00522-6. [DOI] [PubMed] [Google Scholar]
- 118.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG, et al. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophr Res. 2002;58:11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- 120.Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 121.Bousman CA, Chana G, Glatt SJ, Chandler SD, Lucero GR, Tatro E, et al. Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: convergent pathway analysis findings from two independent samples. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:494–502. doi: 10.1002/ajmg.b.31006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rubio MD, Wood K, Haroutunian V, Meador-Woodruff JH. Dysfunction of the ubiquitin proteasome and ubiquitin-like systems in schizophrenia. Neuropsychopharmacology. 2013;38:1910–1920. doi: 10.1038/npp.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arion D, Corradi JP, Tang S, Datta D, Boothe F, He A, et al. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol Psychiatry. 2015;20:1397–1405. doi: 10.1038/mp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hertzberg L, Maggio N, Muler I, Yitzhaky A, Majer M, Haroutunian V, et al. Comprehensive gene expression analysis detects global reduction of proteasome subunits in schizophrenia. Schizophr Bull. 2021;47:785–795. doi: 10.1093/schbul/sbaa160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bousman CA, Luza S, Mancuso SG, Kang D, Opazo CM, Mostaid MS, et al. Elevated ubiquitinated proteins in brain and blood of individuals with schizophrenia. Sci Rep. 2019;9:2307. doi: 10.1038/s41598-019-38490-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nucifora LG, MacDonald ML, Lee BJ, Peters ME, Norris AL, Orsburn BC, et al. Increased protein insolubility in brains from a subset of patients with schizophrenia. Am J Psychiatry. 2019;176:730–743. doi: 10.1176/appi.ajp.2019.18070864. [DOI] [PubMed] [Google Scholar]
- 127.Karanikas E, Daskalakis NP, Agorastos A. Oxidative dysregulation in early life stress and posttraumatic stress disorder: a comprehensive review. Brain Sci. 2021;11:723. doi: 10.3390/brainsci11060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Benzie IF. Lipid peroxidation: a review of causes, consequences, measurement and dietary influences. Int J Food Sci Nutr. 1996;47:233–261. doi: 10.3109/09637489609012586. [DOI] [PubMed] [Google Scholar]
- 129.Finkel T. Oxygen radicals and signaling. Curr Opin Cell Biol. 1998;10:248–253. doi: 10.1016/S0955-0674(98)80147-6. [DOI] [PubMed] [Google Scholar]
- 130.Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–L1028. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
- 131.Higuchi Y. Chromosomal DNA fragmentation in apoptosis and necrosis induced by oxidative stress. Biochem Pharmacol. 2003;66:1527–1535. doi: 10.1016/s0006-2952(03)00508-2. [DOI] [PubMed] [Google Scholar]
- 132.Haines DD, Juhasz B, Tosaki A. Management of multicellular senescence and oxidative stress. J Cell Mol Med. 2013;17:936–957. doi: 10.1111/jcmm.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sharifi-Rad M, Anil Kumar NV, Zucca P, Varoni EM, Dini L, Panzarini E, et al. Lifestyle, oxidative stress, and antioxidants: back and forth in the pathophysiology of chronic diseases. Front Physiol. 2020 doi: 10.3389/fphys.2020.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Miller MW, Sadeh N. Traumatic stress, oxidative stress and posttraumatic stress disorder: neurodegeneration and the accelerated-aging hypothesis. Mol Psychiatry. 2014;19:1156–1162. doi: 10.1038/mp.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miller MW, Lin AP, Wolf EJ, Miller DR. Oxidative stress, inflammation, and neuroprogression in chronic PTSD. Harv Rev Psychiatry. 2018;26:57–69. doi: 10.1097/HRP.0000000000000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tezcan E, Atmaca M, Kuloglu M, Ustundag B. Free radicals in patients with post-traumatic stress disorder. Eur Arch Psychiatry Clin Neurosci. 2003;253:89–91. doi: 10.1007/s00406-003-0413-x. [DOI] [PubMed] [Google Scholar]
- 137.Borovac Štefanović L, Kalinić D, Mimica N, Beer Ljubić B, Aladrović J, Mandelsamen Perica M, et al. Oxidative status and the severity of clinical symptoms in patients with post-traumatic stress disorder. Ann Clin Biochem. 2015;52:95–104. doi: 10.1177/0004563214528882. [DOI] [PubMed] [Google Scholar]
- 138.Zieker J, Zieker D, Jatzko A, Dietzsch J, Nieselt K, Schmitt A, et al. Differential gene expression in peripheral blood of patients suffering from post-traumatic stress disorder. Mol Psychiatry. 2007;12:116–118. doi: 10.1038/sj.mp.4001905. [DOI] [PubMed] [Google Scholar]
- 139.Mokuno K, Ohtani K, Suzumura A, Kiyosawa K, Hirose Y, Kawai K, et al. Induction of manganese superoxide dismutase by cytokines and lipopolysaccharide in cultured mouse astrocytes. J Neurochem. 1994;63:612–616. doi: 10.1046/j.1471-4159.1994.63020612.x. [DOI] [PubMed] [Google Scholar]
- 140.Hu S, Peterson PK, Chao CC. Kappa-opioid modulation of human microglial cell superoxide anion generation. Biochem Pharmacol. 1998;56:285–288. doi: 10.1016/s0006-2952(98)00162-2. [DOI] [PubMed] [Google Scholar]
- 141.Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- 142.Jasinska AJ, Service S, Choi O, De Young J, Grujic O, Kong S, et al. Identification of brain transcriptional variation reproduced in peripheral blood: an approach for mapping brain expression traits. Hum Mol Genet. 2009;18:4415–27. doi: 10.1093/hmg/ddp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cai C, Langfelder P, Fuller TF, Oldham MC, Luo R, van den Berg LH, et al. Is human blood a good surrogate for brain tissue in transcriptional studies? BMC Genomics. 2010;11:589. doi: 10.1186/1471-2164-11-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bergon A, Belzeaux R, Comte M, Pelletier F, Hervé M, Gardiner EJ, et al. CX3CR1 is dysregulated in blood and brain from schizophrenia patients. Schizophr Res. 2015;168:434–443. doi: 10.1016/j.schres.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 145.Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–5. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hess JL, Tylee DS, Barve R, de Jong S, Ophoff RA, Kumarasinghe N, et al. Transcriptome-wide mega-analyses reveal joint dysregulation of immunologic genes and transcription regulators in brain and blood in schizophrenia. Schizophr Res. 2016;176:114–124. doi: 10.1016/j.schres.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are not publicly available due to lack of participant informed consent and ethics approval to deposit the data in a public repository, but are available from the corresponding author on reasonable request.
The code generated during the current study is available from the corresponding author on request.