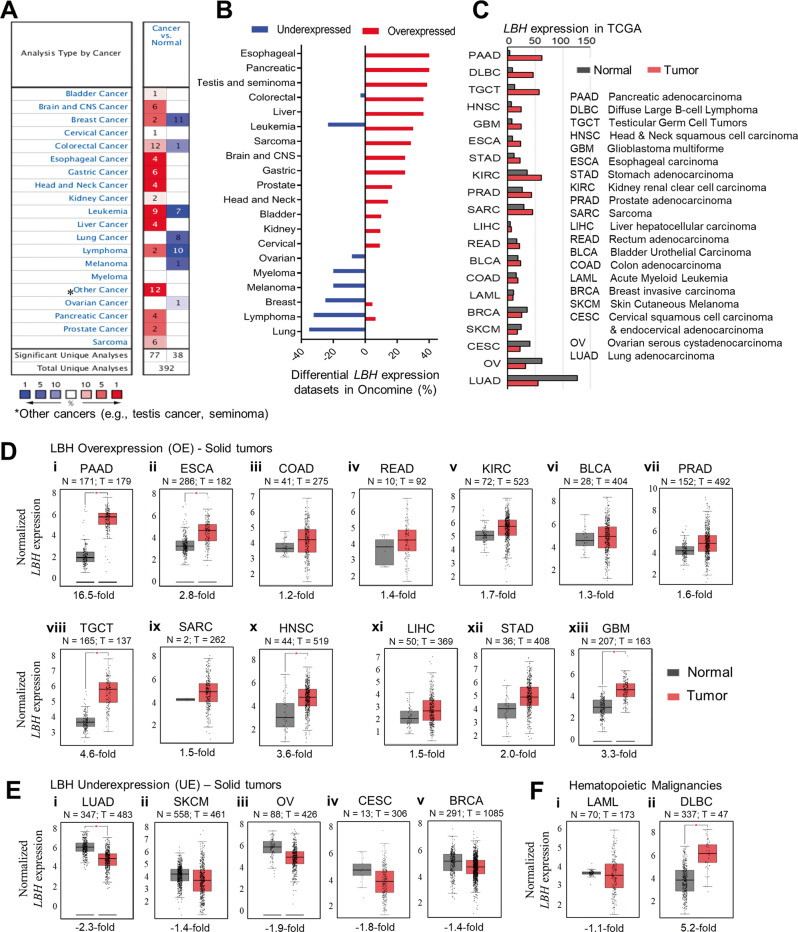
Fig. 1. Meta-analysis of LBH expression in different human cancers.
A The differential expression of LBH mRNA among various cancer types compared to the corresponding normal tissue was generated using Oncomine. The number of datasets with statistically significant (p < 0.05; >1.5-fold change) LBH overexpression (red) or underexpression (blue) are shown. The color scale at the bottom represents the percentages of LBH gene expression ranking in a specific cancer type compared to normal tissue. Dark red, red, and pink indicate that LBH was among the top 1%, top 5%, or top 10%, respectively, of upregulated genes in a dataset. Dark blue, blue, and light blue indicate that LBH was among the top 1%, top 5%, or top 10% of downregulated genes. White indicates no significant changes in LBH gene expression levels. B The percentages of overexpressed and underexpressed analyses for LBH in Oncomine were calculated and plotted in the order of differential LBH expression percentage. C Column plot showing LBH mRNA levels in tumor versus normal tissue in different cancer types in The Consortium Genome Atlas (TCGA) database in descending order of LBH expression (fold change). The abbreviation for each cancer type is explained in the table on the right. Pancreatic cancer (PAAD) showed the highest, and lung cancer (LUAD) the lowest LBH expression compared to normal tissues. D–F Box plots generated from TCGA data (in C) showing the fold changes of LBH mRNA in tumor (red) versus normal tissues (gray). D Solid tumors with LBH overexpression; E solid tumors with LBH underexpression; and F blood cancers with deregulated LBH expression. The threshold was set at p-value = 0.05. The number of normal (N) and tumor (T) tissues is indicated for each cancer type. LBH expression data were extracted from the TCGA and GTEx databases using GEPIA2. See also Fig. S1.