Figure 3.
HERC2 recognizes chaperonin subunits using the adaptor ZNRD2
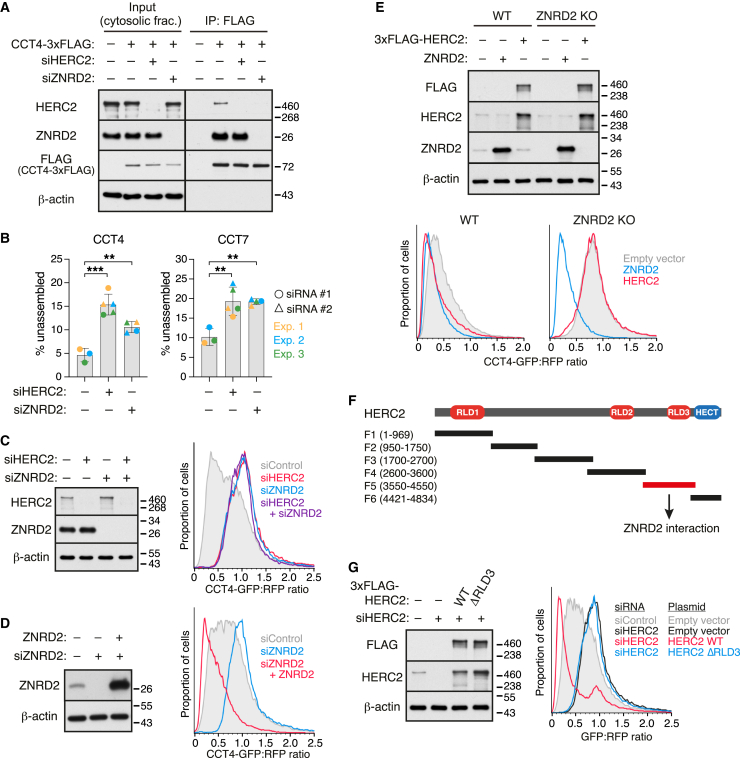
(A) Cells treated with siRNAs against HERC2 or ZNRD2 were transfected with CCT4-3xFLAG, followed by non-denaturing IP using anti-FLAG affinity resin. Input and IP samples were analyzed by immunoblotting for the indicated proteins.
(B) Cytosolic extracts from cells treated with siRNAs targeting HERC2, ZNRD2, or control were fractionated by sucrose gradient centrifugation to quantify the proportion of unassembled CCT4 and CCT7 subunits (see Figures S4A and S4B). Data are mean ± SD of data points (dots) from three independent experiments (color-coded). ∗∗p < 0.01 and ∗∗∗p < 0.001 vs. control siRNA by one-way ANOVA with Dunn’s multiple comparisons test.
(C) Cells treated with the indicated siRNAs were transfected with the dual-color CCT4 reporter and analyzed by immunoblotting (left) and flow cytometry (right).
(D) Cells treated with siRNAs against ZNRD2 were transfected with the CCT4 reporter and siRNA-resistant ZNRD2, followed by immunoblotting (left) and flow cytometry (right).
(E) Wild-type (WT) and ZNRD2-knockout (KO) Flp-In T-REx 293 cells were transfected with the dual-color CCT4 reporter and either 3xFLAG-HERC2, ZNRD2, or empty vector. Cells were analyzed by immunoblotting (top) and flow cytometry (bottom).
(F) Diagram illustrating the domain structure of human HERC2 (4,834 amino acids) and overlapping HERC2 fragments tested for interaction with ZNRD2 in Figure S4G. HERC2 contains three RCC1-like domains (RLD1–3) and a C-terminal HECT E3 ligase catalytic domain (HECT). The HERC2 fragment containing RLD3 (F5; highlighted in red) interacts with ZNRD2 (see Figure S4G).
(G) Cells treated with HERC2-targeting siRNA were transfected with the CCT4 reporter together with empty vector, siRNA-resistant 3xFLAG-HERC2 (WT), or its deletion mutant lacking RLD3 (ΔRLD3). Cells were analyzed by immunoblotting (left) and flow cytometry (right).
See also Figure S4.