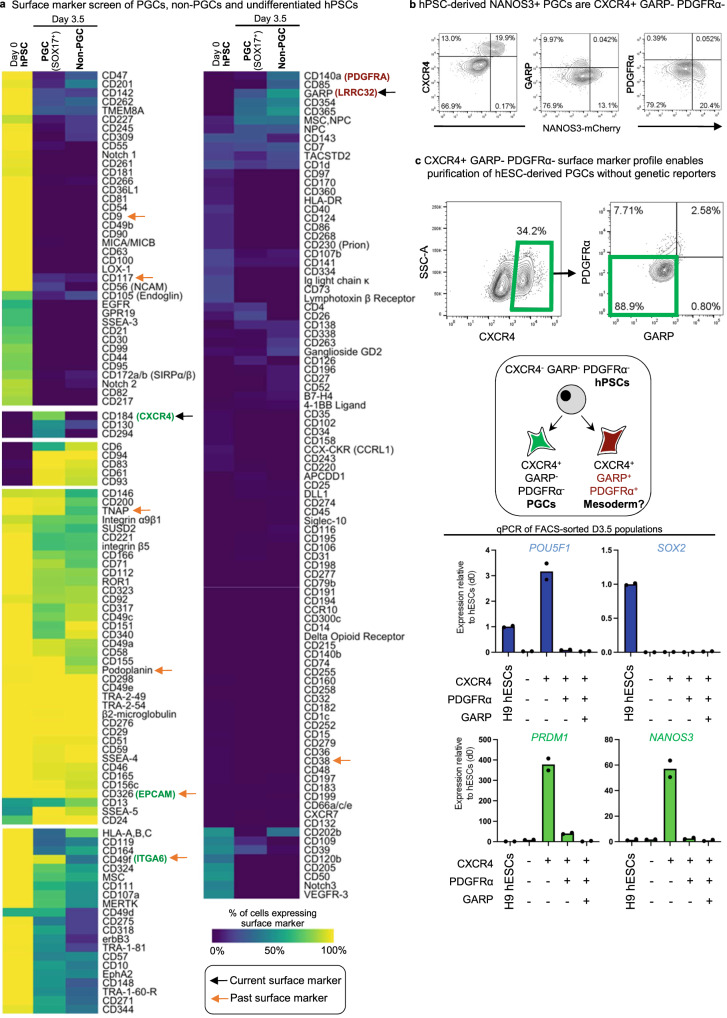
Fig. 5. High-throughput screening identifies a CXCR4+ PDGFRα− GARP− cell-surface marker signature for hPSC-derived PGCLCs.
a Heatmap of surface markers expressed in undifferentiated hPSC (D0), D3.5 SOX17-GFP+ PGCLCs, and D3.5 SOX17-GFP− non-PGCLCs identified from LEGENDScreen; to discriminate PGCLCs vs. non-PGCLCs, SOX17-GFP hESCs were differentiated for D3.5 and then subgated on GFP+ and GFP− before further analysis of surface marker expression; color shades represent the percentage of cells in each expression that are positive for a given marker; each row depicts expression of a single surface marker across all populations. b Flow cytometry analysis of D3.5 differentiated NANOS3-mCherry hESCs reveals CXCR4, GARP, and PDGFRα expression relative to NANOS3-mCherry fluorescent reporter expression. c Flow cytometry gating strategy to identify CXCR4+/GARP−/PDGFRα− PGCLCs derived from H9 hESCs (that did not carry any fluorescent reporters) that were differentiated for D3.5; various cell populations from the D3.5 population were FACS sorted and subject to qPCR analysis, revealing that pluripotency and PGC markers are restricted to the CXCR4+/GARP−/ PDGFRα− subset and therefore reaffirming its PGCLC identity. N = 2 biological replicates, Data are presented as mean values. Source data are provided as a Source Data file.