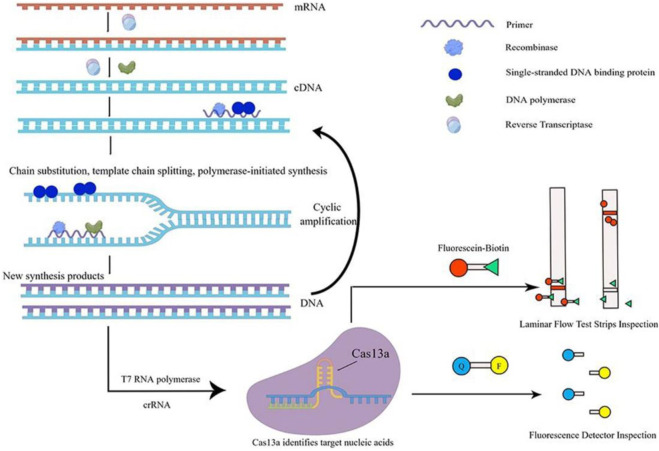
FIGURE 2.
Schematic diagram of single-tube one-step method. This figure was drawn by Figdraw. The specific principle of single-tube one-step detection of RNA viruses is as follows: in the nucleic acid amplification system under constant temperature conditions, firstly, dNTP is used as substrate material, mRNA is used as a template, and under the action of reverse transcriptase, a cDNA single-strand complementary to the RNA template is synthesized to form DNA-RNA heterozygote. In this DNA-RNA heterozygote, RNA is specifically degraded by reverse transcriptase, and then dNTP is used as the substrate, the first strand of cDNA is used as a template, and under the action of DNA polymerase, the second strand of cDNA is synthesized, and finally the double-stranded DNA molecule is formed, that is, the DNA synthesis process guided by RNA is completed. Then, this double-stranded DNA molecule is used as the template DNA. When the primer finds the complementary sequence that perfectly matches the template DNA, with the help of recombinase, the double-stranded structure of the template DNA is opened, while SSB stabilizes the displaced DNA chain, and the extension of the strand is completed under the action of DNA polymerase to form a new complementary strand of DNA. The T7 RNA polymerase transcribed the amplified product into RNA, while the Cas13a protein specifically bound the target nucleic acid under the guidance of crRNA, the protein structure was changed, and was converted into ribonuclease, which was non-specific to cut RNA. The incidental cutting activity could cut the introduced externally derived FQ-coated ssRNA probe. The fluorescence signal can be amplified rapidly to detect the target nucleic acid quickly.