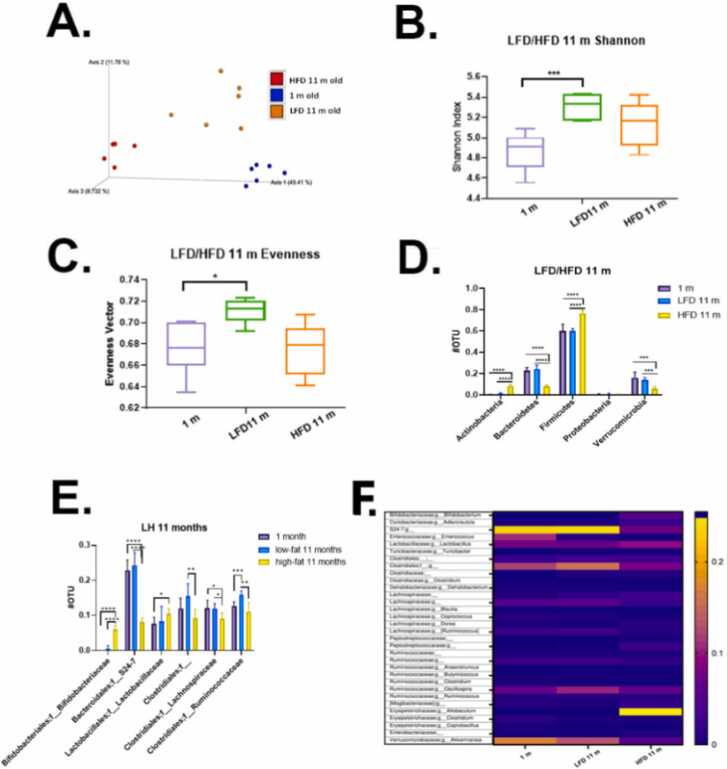
Fig. 6.
Microbial communities’ analysis: age and LFD vs HFD. A. Phylogenetic based beta diversity-principal component analysis (PCA) plot of fecal microbiota was examined. Plots based on unweighted UniFrac distance matrices of microbial communities in fecal samples, three separated clusters were displayed. PC1 (x‐axis) explained 49.41 %, PC2 (y‐axis) explained 11.78 % of variability, PC3 (z‐axis) explained 8.73 % of the variability. B. Phylogenetic based alpha diversity-the Shannon index. Lower Shannon vectors indicate the lower diversity of the microbial communities. LFD mice had the highest diversity than the other two groups. 1-month old mice majority microbial communities came from the mother and less diverse and richness influenced by diet. Microbiota in young mice tend to be less diverse than that of older mice. C. Alpha diversity-the evenness was compared in three groups. The LFD group had the highest richness and evenness than the other two groups, indicating LFD fed mice microbiome community has a small disparity between the number of individuals within each species. D. Quantitative phyla levels of operational taxonomic unit (OTU) was compared. E. Family level of operational taxonomic unit (OTU) was compared. The HFD mice samples increased in Bifidobacteriaceae, Lactobacillaceae; decreased in S24–7, Clostridiales, Lachnospiraceae, Ruminococcaceae and one undefined family. Significance, *p < 0.05, * *p < 0.01, * **p < 0.001, * ** * p < 0.0001. F. Microbial communities’ analysis. Heatmap plots of 11 months mice fecal samples showed the gut microbiota composition between samples at the genus level. Relative abundances of individual taxa (rows) in each sample (columns) are indicated in the associated color scale.