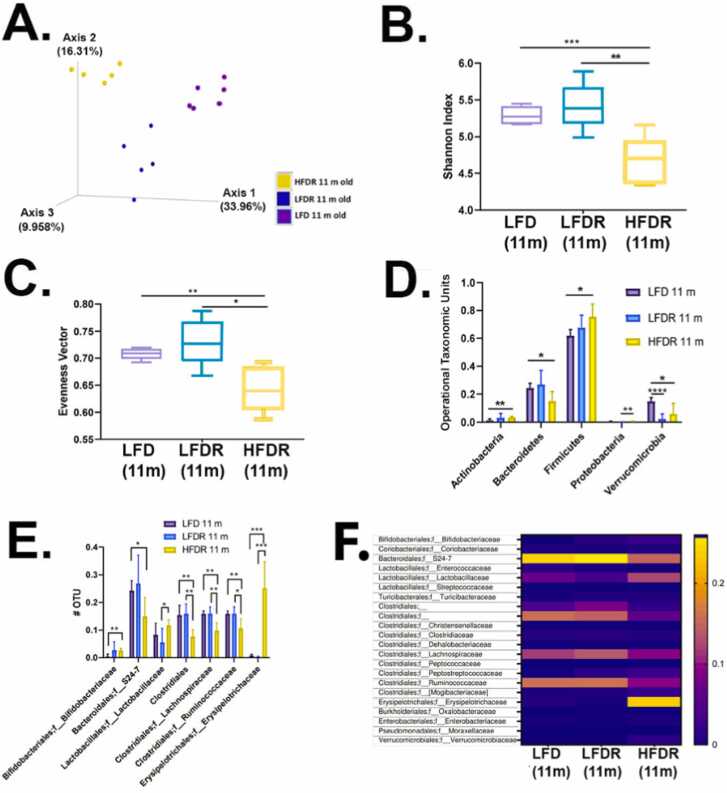
Fig. 7.
Microbial diversity of after 10 months exposure to Red 40. A. Phylogenetic based beta diversity, a qualitative principal component analysis (PCA) plot of fecal microbiota. Compare LFD 11-month old mice fecal samples, LFD/HFD with Red 40 11 months old mice fecal sample. Unweighted UniFrac distance matrices of microbial communities in fecal samples, PC1 (x‐axis) explained 33.96 %, PC2 (y‐axis) explained 16.31 % of variability, PC3 (z‐axis) explained 9.96 % of the variability. B. Phylogenetic based alpha diversity, the Shannon index. Lower Shannon vectors indicate the lower diversity of the microbial communities. Comparing HFD with Red 40 11 months old mice to LFD with or without Allura Red AC 11 months old mice Shannon vector significantly decreased. *p < 0.05, * *p < 0.01, * **p < 0.001. C. Comparing the alpha diversity evenness. Comparing the HFD with Red 40 11 months old mice to LFD with or without Red 40 11 months old mice samples, diversity of evenness significantly decreased. Significant, D. Operational taxonomic unit (OTU) of the Phyla level were compared: LFD and HFD with and without Red 40 11 months mice microbiota. The significantly increased in Actinobacteria, Firmicutes; and Proteobacteria. Decreased in Bacteroidetes only in HFD with Red 40 samples, and decreased Verrucomicrobia in both LFD and HFD with Red 40 samples. E. Comparing the OTU of the family level: Increased in Bifidobacteriaceae and Erysipelotrichaceae, especially in overlaid with HFD and Red 40, decreased in Bacteroidales family level in S24–7, especially in overlaid with HFD and Red 40; Clostridiales family level in Ruminococcaceae and Lachnospiraceae, especially in overlaid with HFD and Red 40. F. Heatmap of family level. Heatmap plots show the gut microbiota composition between samples at the family level. Relative abundances of individual taxa (rows) in each sample (columns) are indicated in the associated color scale. *p < 0.05, * *p < 0.01, * **p < 0.001, * ** * p < 0.0001.