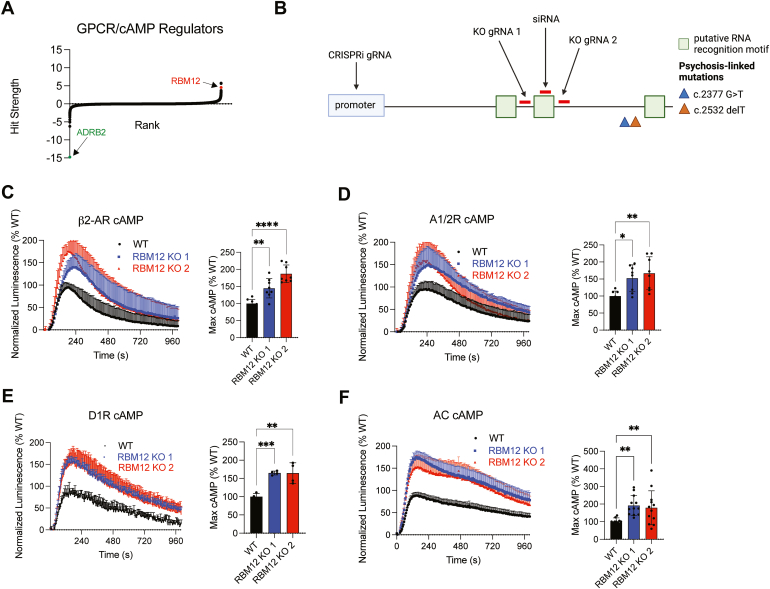
Figure 1.
RBM12 loss leads to hyperactive GPCR/cAMP signaling.A, genes (20,528) ordered by hit strength (product of phenotype score and -log10(p-value)). Candidate repressors exhibit hit strength > 0, and candidate activators exhibit hit strength < 0 in a genome-wide CRISPR screen for GPCR/cAMP regulators (8). B, schematic of multiple strategies to deplete RBM12 in HEK293 using CRISPRi, RNAi, and CRISPR knockout, depicting positions of guide RNAs, siRNA, and RBM12 SNPs (c.2377G>T and c.2532delT) implicated in psychosis. C, luminescent GloSensor measurement of cAMP accumulation in response to β2-AR agonist isoproterenol (Iso), 100 nM (n = 8). D, luminescent GloSensor measurement of cAMP accumulation in response to activation of A1/2R with 10 μM NECA (n = 8). E, luminescent GloSensor measurement of cAMP accumulation in response to D1R-selective agonist SKF-81297, 10 nM (n = 4). F, luminescent GloSensor measurement of cAMP accumulation in response to direct adenylyl cyclase activation with forskolin, 1 μM (n = 12). All data are mean ± SD. Statistical significance was determined using one-way ANOVA with Dunnett’s correction (C–F). See also Fig. S1. ∗∗∗∗ = p < 0.0001, ∗∗∗ = p < 0.001, ∗∗ = p < 0.01, ∗ = p < 0.05. β2-AR, beta-2-adrenergic receptor; CRISPRi, CRISPR interference; GPCR, G protein–coupled receptor; NECA, 5′-(N-Ethyl Carboxamide) adenosine; RBM, RNA-binding motif.