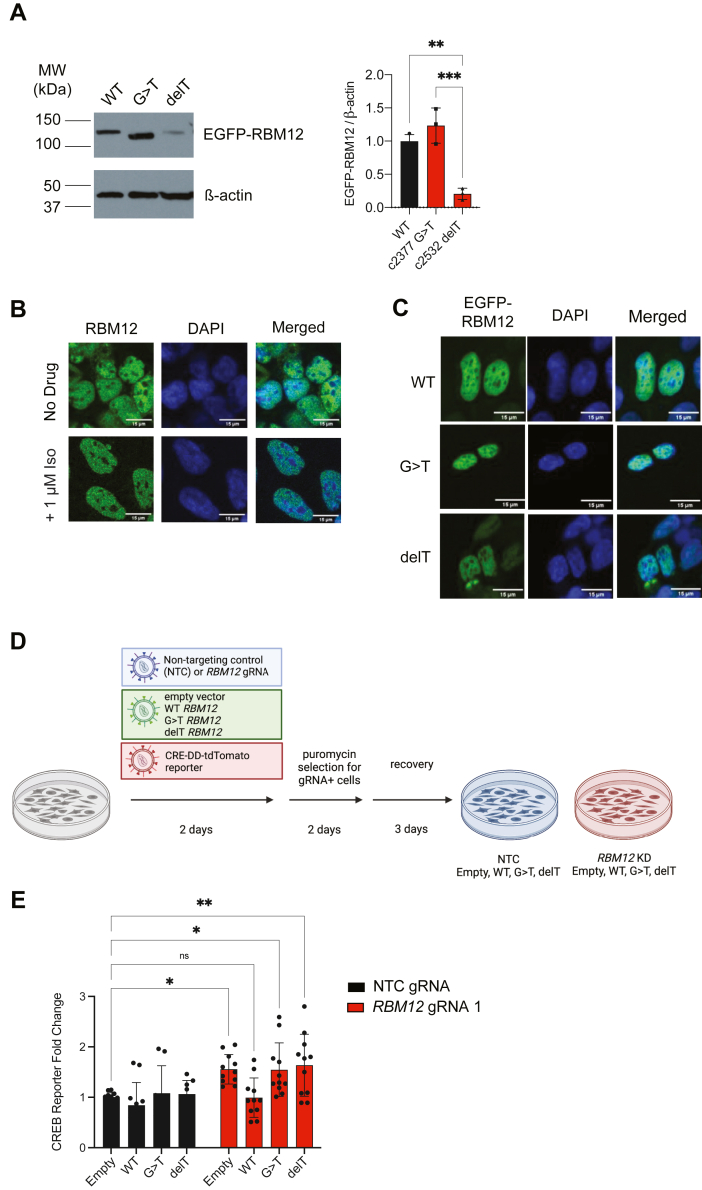
Figure 4.
Expression of disease-associated variants in RBM12 knockdown cells does not rescue the hyperactive GPCR-dependent transcriptional signaling.A, Western blot analysis of EGFP-tagged WT, c.2377G>T and c.2532delT RBM12 (n = 3) probed with antibody recognizing EGFP. All data are normalized relative to WT. See also Fig. S7. B, fixed cell fluorescence microscopy analysis of endogenous RBM12 in untreated and stimulated (1 μM Iso for 20 min) HEK293 cells. C, localization of EGFP-tagged WT, c.2377G>T or c.2532delT RBM12 by fluorescence microscopy. D, schematic of the flow cytometry–based rescue experiment. E, flow cytometry measurement of the fluorescent CREB transcriptional reporter (CRE-DD-tdTomato) in response to 1 μM Iso and 1 μM Shield for 6 h (n = 8–11). Data are normalized relative to the “NTC + empty vector” sample values. All data are mean ± SD. Statistical significance was determined using one-way ANOVA (A) or two-way ANOVA with Dunnett’s correction (E). See also Fig. S4. ∗∗∗ = p < 0.001, ∗∗ = p < 0.01, ∗ = p < 0.05. GPCR, G protein–coupled receptor; NTC, nontargeting control; RBM, RNA-binding motif.