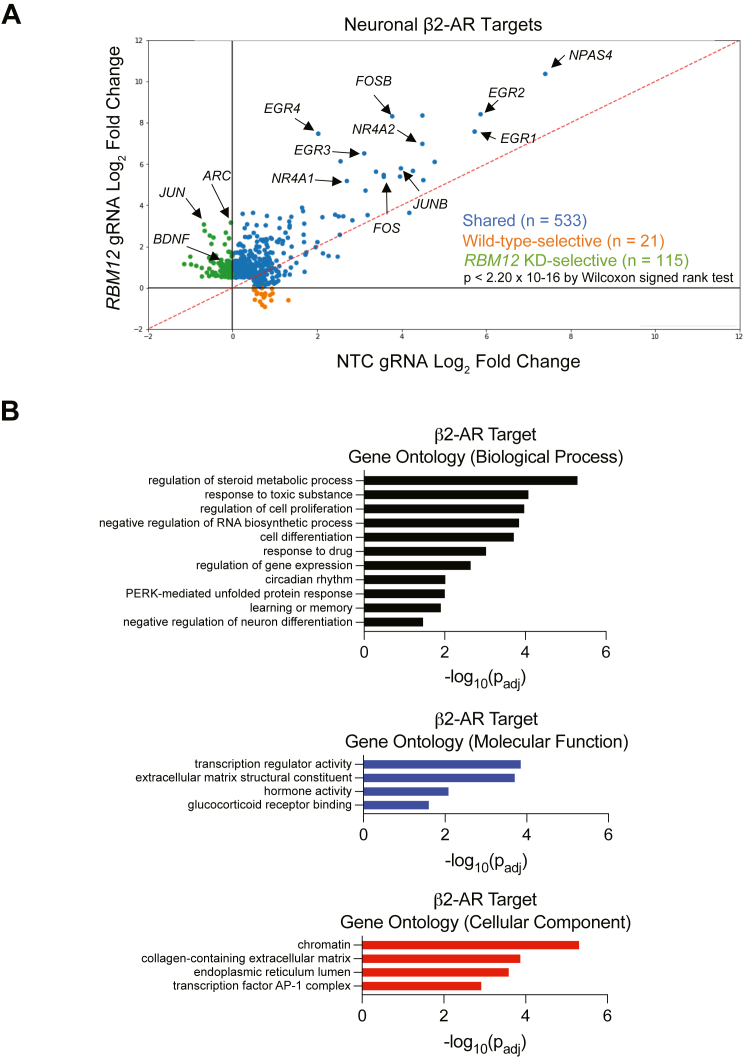
Figure 6.
RBM12 loss impacts the β2-AR neuronal transcriptional responses. A, scatter plot showing gene fold induction (log2 Iso/no drug) of neuronal β2-AR targets (n = 669) identified by RNA-seq analysis (n = 3 per cell line per drug condition). Blue dots represent genes that were induced by a 1-h treatment with 1 μM Iso in both WT (NTC gRNA) and RBM12 KD (RBM12 gRNA) neurons. Orange dots represent genes that were induced only in WT and unchanged or downregulated in RBM12 KD neurons. Green dots represent genes that were induced only in RBM12 KD neurons and unchanged or downregulated in WT. Indicated by arrows are a subset of genes with established roles in neuronal activity. The underlying information is summarized in Table S1. B, gene ontology categories enriched among the neuronal β2-AR targets from (A). Statistical significance was determined using Wilcoxon signed rank test (A) and Fisher’s exact test (B). See also Fig. S6. β2-AR, beta-2-adrenergic receptor; NTC, nontargeting control; RBM, RNA-binding motif.