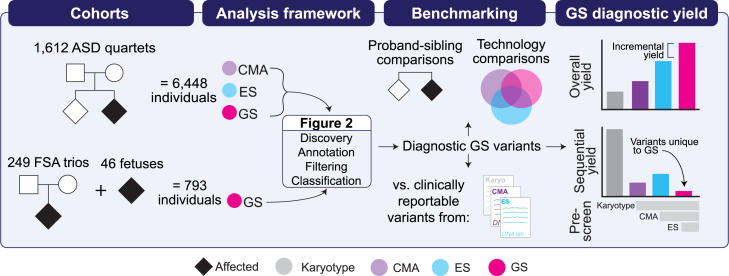
Figure 1.
Overall study design
We performed genome sequencing (GS) on 7,241 individuals from two phenotypically ascertained cohorts: autism spectrum disorder (ASD) and fetal structural anomalies (FSAs). The ASD quartet families (n = 6,448 individuals) included one affected proband with ASD, one unaffected sibling, and two unaffected parents. The prenatal cohort included 249 trios (n = 747 individuals) comprising a fetus with an FSA detected by ultrasound and two unaffected parents as well as 46 singleton fetuses ascertained for a diagnostic procedure performed in pregnancy. Fetuses from the 249 trios were pre-screened with one or more standard-of-care diagnostic tests (karyotype, chromosomal microarray [CMA], and/or exome sequencing [ES]) and the 46 singleton fetuses were pre-selected on the basis of having a clinically reportable variant identified by one of the same three standard-of-care tests. For the 1,612 ASD quartet families, we had access to unfiltered data from CMA, ES, and GS available for analysis (see subjects and methods for more details). We performed multiple benchmarking analyses, including comparing the yield of diagnostic variants between ASD probands and their unaffected siblings, direct technology comparisons in the ASD probands, and comparisons against results from clinical diagnostic tests in the fetuses. We assessed the performance of GS by considering the overall, incremental, and sequential diagnostic yields provided by this technology. Plots are demonstrative only and are not drawn to scale nor reflective of real data.