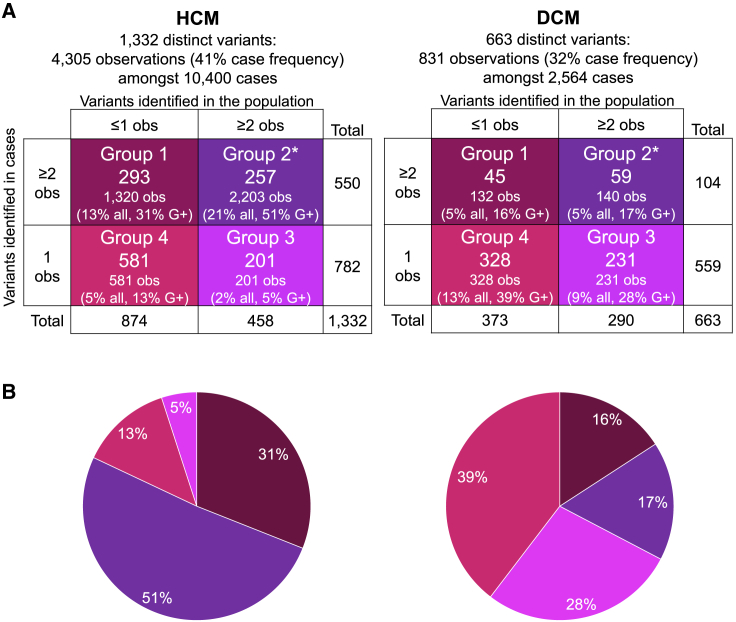
Figure 3.
Penetrance of individual variants could be estimated for 316 recurrently observed rare variants from group 2
(A) The figure shows variant counts and subgroups for rare variants in HCM-associated (left) and DCM-associated (right) genes. (B) The pie charts plot the proportion of all variant observations in each subgroup (also denoted as “G+”). The observations approximate to the number of individuals with variants, although a small number of individuals may carry more than one variant. All, denotes frequency of the variant in affected individuals; obs, denotes observations of allele count. Group 1: variants observed recurrently in affected individuals and absent or singleton in the population; penetrance estimates are unreliable as the population frequency is uncertain. This group is expected to include most definitively pathogenic, high-penetrance variants. Group 2∗: variants observed recurrently in affected individuals and the wider population; these are the variants most likely to be observed as secondary findings. ∗Penetrance can be estimated. Group 3: variants observed once in affected individuals and recurrently in the population; penetrance estimates are unreliable, as the case frequency is uncertain. Variants in this group are likely either not pathogenic or have low penetrance. Group 4: variants are singleton in affected individuals and absent or singleton in the population; current data is too sparse to estimate penetrance.