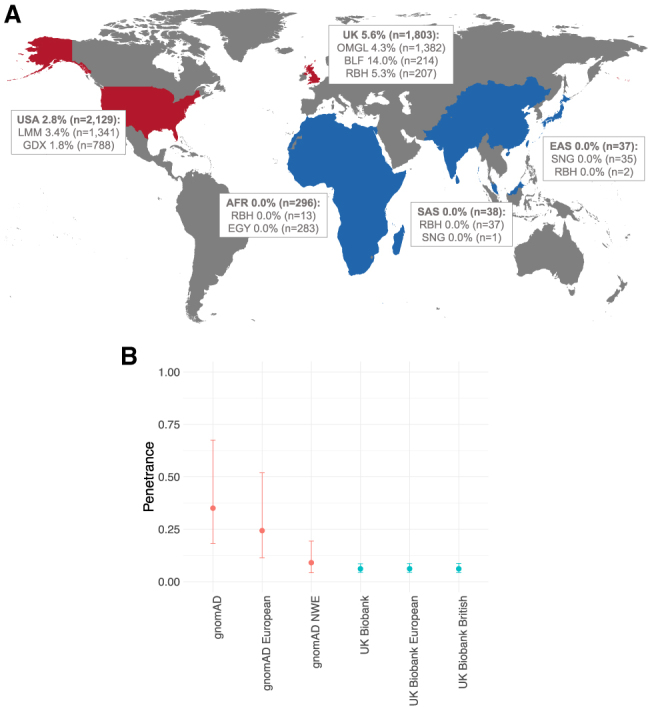
Penetrance estimates are inflated with underestimated population frequency
(A) The map of the world emphasizes the large proportion of observations of MYBPC3 c.1504C>T (p.Arg502Trp) in HCM-affected individuals of Northwestern European (NWE) ancestry. The numbers on the map are the counts of rare-variant-genotype-positive observations (n ≈ cohort participants) from each cohort with the specified ancestry, and the percentages derive the proportion of observations that are due to the MYBPC3 c.1504C>T (p.Arg502Trp) variant. (B) The graph shows the estimated penetrance and 95% confidence interval for the variant on the basis of subgroups of reference dataset participants included. The penetrance is inflated when estimated with gnomAD because the variant is most common in participants with NWE ancestry (which dominates the UKBB dataset). Population frequency of gnomAD, UK Biobank, and Ensembl population genetics showed that this variant (rs375882485) is only found multiple times in NWE ancestry sub-cohorts. The map excludes Antarctica for figure clarity. A limitation is the low sample sizes for AFR, SAS, and EAS ancestries.26
