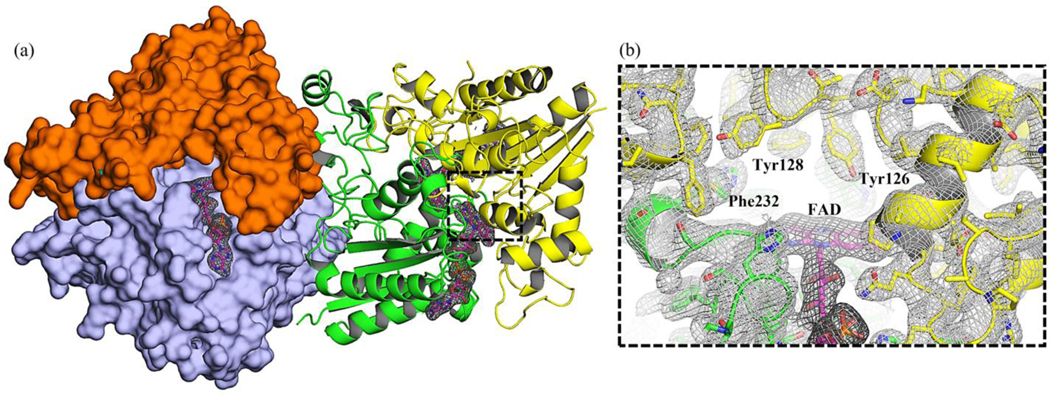
Figure 6:
SFX structure of human NQO1 obtained at MFX. (a) The two dimers of NQO1 in the asymmetric unit are depicted. The individual monomers are highlighted in green, yellow, orange, and light blue. The cofactor FAD is shown as pink sticks. The catalytic site of one of the monomers is black-boxed. (b) Closer view of the catalytic site in (a). The electron 2mFc-DFo density maps at the catalytic site contoured at 1σ are shown. Residues Tyr126, Tyr128, and Phe232 which are key in the function of the enzyme are highlighted. Our results also suggest the presence of different conformational sub-states prior to NAD(P)H binding and consequent flavin reduction, thus supporting a conformational selection mechanism and providing structure-function information at high resolution.