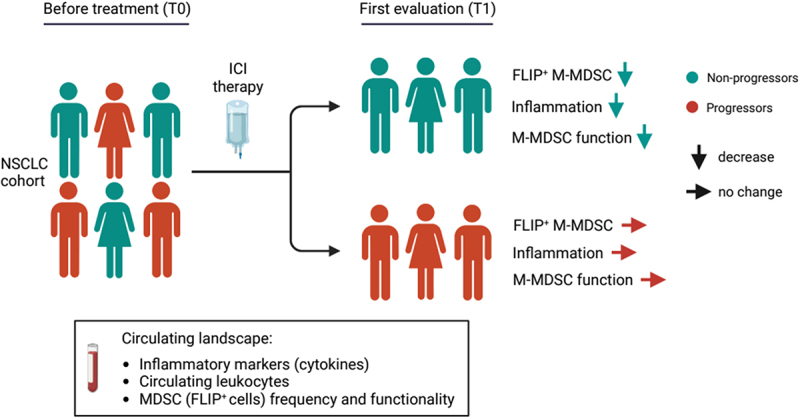
ABSTRACT
Cancer cells favor the generation of myeloid cells with immunosuppressive and inflammatory features, including myeloid-derived suppressor cells (MDSCs), which support tumor progression. The anti-apoptotic molecule, cellular FLICE (FADD-like interleukin-1β-converting enzyme)-inhibitory protein (c-FLIP), which acts as an important modulator of caspase-8, is required for the development and function of monocytic (M)-MDSCs. Here, we assessed the effect of immune checkpoint inhibitor (ICI) therapy on systemic immunological landscape, including FLIP-expressing MDSCs, in non-small cell lung cancer (NSCLC) patients. Longitudinal changes in peripheral immunological parameters were correlated with patients’ outcome. In detail, 34 NSCLC patients were enrolled and classified as progressors (P) or non-progressors (NP), according to the RECIST evaluation. We demonstrated a reduction in pro-inflammatory cytokines such as IL-8, IL-6, and IL-1β in only NP patients after ICI treatment. Moreover, using t-distributed stochastic neighbor embedding (t-SNE) and cluster analysis, we characterized in NP patients a significant increase in the amount of lymphocytes and a slight contraction of myeloid cells such as neutrophils and monocytes. Despite this moderate ICI-associated alteration in myeloid cells, we identified a distinctive reduction of c-FLIP expression in M-MDSCs from NP patients concurrently with the first clinical evaluation (T1), even though NP and P patients showed the same level of expression at baseline (T0). In agreement with the c-FLIP expression, monocytes isolated from both P and NP patients displayed similar immunosuppressive functions at T0; however, this pro-tumor activity was negatively influenced at T1 in the NP patient cohort exclusively. Hence, ICI therapy can mitigate systemic inflammation and impair MDSC-dependent immunosuppression.
KEYWORDS: NSCLC (non-small cell lung cancer); M-MDSCs (monocytic myeloid-derived suppressor cells), c-FLIP (cellular FLICE [FADD-like IL-1β-converting enzyme]- inhibitory protein); ICI (immune checkpoint inhibitor)
GRAPHICAL ABSTRACT
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide and is responsible for more than 1.7 million deaths per year (http://gco.iarc.fr). Although histopathological features remain the standard basis for diagnosis, recent advances in next-generation sequencing and high-throughput analyses revealed a multifaceted tumor framework based on the intrinsic properties of both cancer cells, such as genetic mutations and genomic heterogeneity,1–3 and cancer cell-extrinsic features, such as immune cells located in the tumor microenvironment (TME), which influence tumorigenesis, tumor progression, and response to therapy.4–6 Therefore, non-small cell lung cancer (NSCLC) is recognized as a heterogeneous set of diseases that can benefit from innovative approaches, such as targeted therapies7–9 and immunotherapy.10–13
The discovery of programmed cell death 1 (PD-1)/PD-1 ligand 1 (PD-L1) immune checkpoint inhibitors (ICI) marks the beginning of a new era in the treatment of NSCLC.14 A pioneering study demonstrated the efficacy and durable response of immunotherapy for the treatment of advanced NSCLC, also suggesting a correlation between PD-L1 expression on tumor cells and objective response.15 Currently, immunotherapy as a single agent or in combination with chemotherapy is the standard first-line approach in patients with non-oncogene-addicted stage IV NSCLC.11 Despite reproducible positive results, only a minority (<20%) of the patients show long-term benefit from ICI immunotherapy and most will progress at any time during treatment; furthermore, a non-negligible proportion of patients receiving ICI do not respond to treatment despite high PD-L1 expression. Several mechanisms of resistance to ICIs have been described, including changes in the cell composition of the TME and immune response.16–18 Therefore, new predictive biomarkers for immunotherapy resistance are important to maximize therapeutic effectiveness and test new treatment options, including combined strategies.
Tumor-promoting inflammation and evasion of immune destruction are now recognized hallmarks of cancer.19,20 Cancer cells impair the immune response by altering physiological systems such as myelopoiesis to expand cell subsets with pro-tumoral features, such as myeloid-derived suppressor cells (MDSCs).21–23 MDSCs are a heterogeneous population of bone marrow-derived cells containing a mixture of mature myeloid cells and precursors of monocytes and granulocytes that share common suppressive activity in relation to both innate and adaptive immunity.24,25 MDSCs employ different mechanisms to restrain the anti-tumor immune response, including the release of anti-inflammatory cytokines [interleukin (IL)-10 and transforming growth factor (TGF)β], consumption of nutrients in the environment by the over-expression of metabolism-associated enzymes (arginase 1 (Arg1), inducible nitric oxide synthase (iNOS/NOS2), and indoleamine 2,3-dioxygenase 1 (IDO1)), production of both reactive oxygen species (ROS) and reactive nitrogen species (RNS), and the expression of tolerogenic molecules (PD-L1).26–29 Furthermore, MDSCs contribute to non-immunologic aspects of tumor biology, including angiogenesis and metastasis.23,30 The clinical significance of MDSCs has been validated by numerous studies, highlighting the correlation between circulating and intra-tumoral MDSC frequencies with tumor stage progression and treatment resistance.31–34 The anti-apoptotic molecule, cellular FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein (c-FLIP), was proposed as a driver for the development of monocytic (M)-MDSCs in preclinical cancer models.35 We demonstrated that c-FLIP directly regulates the tolerogenic properties of human monocytes, in part through activation of the canonical nuclear factor-κB (NF-κB) signaling pathway; furthermore, the presence of immunosuppressive c-FLIP-overexpressing PD-L1+CD14+ cells in combination with high levels of serum IL-6 was an independent, negative prognostic factor for both overall survival (OS) and disease-free survival (DFS) in patients with pancreatic ductal adenocarcinoma (PDAC).36 Consistent with the immune regulatory properties, c-FLIP-expressing monocytes isolated from coronavirus disease (COVID)-19 patients displayed immunosuppressive functions and released high amounts of pro-inflammatory cytokines through the activation of an aberrant FLIP-dependent signal transducer and activator of transcription 3 (STAT3) pathway,37 which controls several MDSC functions.38,39 Collectively, these data emphasize c-FLIP contribution in reprogramming monocytes into MDSCs under different pathological settings, including cancer.
In this longitudinal prospective circumscribed study, we sought to investigate the effect of ICI on circulating immunological landscape, including inflammation-associated soluble factors as well as the immunosuppressive properties of FLIP-expressing M-MDSCs.
Patients and methods
Patients and study approval
Patients with advanced NSCLC treated with ICIs (pembrolizumab, nivolumab, and atezolizumab) as first or second-line treatment were prospectively enrolled between January 2019 and September 2020 at the Oncology Unit of the University Hospital of Verona. We retrospectively defined as: progressor (P) those patients who experienced a disease progression within 6 months from the first ICI administration after at least 6 weeks of treatment, and non-progressor (NP) those patients who experienced progressive disease during treatment after at least 6 months of clinical/radiological benefit.40,41 Blood samples were collected from patients at baseline (before starting ICIs, defined as T0) and after 6 weeks (defined as T1). All the patients provided written informed consent before sampling and for the use of their clinical and biological data. This study was approved by the local Ethical Committee (Prot. 1839 CESC; P.I. Silvia Sartoris) and conducted according to the Declaration of Helsinki and Good Clinical Practice.
Flow cytometry of immune subsets
Whole blood samples (100 μl/tube) were stained for cell-surface markers detection with anti-human HLA-DR-PE, CD14-ECD, CD38-PE-Cy5, CD45-Al700, CD3-A750, CD16-V450, CD4-V500, CD57-FITC, CD8-PE-Cy7, CD14-APC, CD19-V500, all from Beckman Coulter Life Sciences (Brea, CA, USA). Samples were stained by using IMMUNOPREP Reagent kit (Beckman Coulter Life Sciences, Brea, CA, USA) and Workstation PrepPlus 2 (Beckman Coulter Life Sciences, Brea, CA, USA) according to manufacturer’s instructions. 1 × 106 PBMCs were incubated with FcReceptor Blocking reagent (Miltenyi Biotec, Bologna, Italy) and stained with anti-human CD14-APCH7, CD16-FITC, HLA-DR-PE-Cy7, and LIVE/DEAD™ Fixable Aqua Dead Cell Stain (eBioscience, Thermo Fisher Scientific, Waltham, MA, USA). Samples were fixed and permeabilized with Foxp3/Transcription Factor Staining Buffer Set (eBioscience, Thermo Fisher Scientific, Waltham, MA, USA) and incubated with anti-FLIP antibody (D5J1E, Cell Signaling Technologies, Danvers, MA, USA) and PE-F(ab’)2 Donkey anti-Rabbit IgG (BD Biosciences, San Jose, CA, USA). Samples were acquired with FACS Canto II (BD Biosciences, San Jose, CA, USA) and Navios EX (Beckman Coulter Life Sciences, Brea, CA, USA), and analyzed by using FlowJo software (Tree Star, Inc. Ashland, OR, USA) and Navios EX Software (Beckman Coulter Life Sciences, Brea, CA, USA), respectively. Circulating cell count (cells/μl) of specific subsets was obtained by transforming the proportion obtained by flow cytometry data using the white blood cell (WBC) count tested on the same blood samples.
Detection of cytokines
Circulating cytokines in NSCLC patient-derived frozen plasma samples were assessed by automated immunoassay workflow. In detail, IL-1β, IL-8, TNF-α, and IL-6 were quantified by Ella™ technology (Bio-Techne, Minneapolis, MN, USA). GM-CSF, IFN-γ, IL-12p70, IL-13, IL-18, IL-2, IL-5, IL-10, IL-17A, IL-27, IL-1α, IL-15, IL-1RA, IL-7, CCL11, CXCL1, CXCL10, CCL2, CCL3, CCL4, CCL5, and SDF-1a were detected by Luminex Performance Assay 3-plex Kit (R&D System Minneapolis, MN, USA) according to manufacturer’s instructions. TNF-β, IL-31, IFN-α, IL-21, IL-22, IL-23, IL-9, and IL-4 levels were evaluated with the same technology but their concentration resulted below the detection limit for many patients and were consequently excluded from the analysis. Reference cytokine ranges were obtained from the literature.42–51
Cell isolation and culture
Peripheral blood mononuclear cells (PBMCs) were isolated from NSCLC patient-derived blood samples by Ficoll-Hypaque (GE Healthcare, Uppsala, Sweden) gradient centrifugation. PBMCs from healthy donors (HDs) were isolated from leukocyte-enriched buffy coats from healthy volunteers (Transfusion Center, University and Hospital Trust of Verona, Verona, Italy) by using the same methods. CD14+ monocytes were isolated by immunomagnetic sorting (Miltenyi Biotec, Bologna, Italy) according to the manufacturer’s instructions and after checking their purity by flow cytometry they were cryopreserved in liquid-phase nitrogen. PBMCs from HDs were stained with 1 μM of CellTrace Violet (eBioscience, Thermo Fisher Scientific, Waltham, MA, USA) in PBS by 5 min incubation at 37°C, protected from light. Labeled PBMCs were stimulated with 0.6 μg/mL anti-CD3 and 5 μg/mL anti-CD28 (OKT-3 and CD28.2 clone, respectively – both from eBioscience, Thermo Fisher Scientific, Waltham, MA, USA) for 4 days and co-cultured with thawed, NSCLC patient-derived CD14+ cells at 3:1 ratio (CD14+ cells: PBMCs). Cell cultures were incubated at 37°C and 8% CO2 in l-arginine-free-RPMI (Biochrom AG, Berlin, Germany), supplemented with 2 mM L-glutamine (Euroclone, Milano, Italy), 150 μM l-arginine (Sigma-Aldrich, St Louis, MO, USA), 10% FBS (Superior, Merck, Darmstadt, Germany), 10 U/ml penicillin and streptomycin (Euroclone, Milano, Italy), and 0.1 mM HEPES (Euroclone, Milano, Italy). At the end of the co-culture, cells were stained with PE-Cy7 conjugated anti-CD3 (eBioscence, Thermo Fisher Scientific) and acquired with a FACSCanto II (BD, Franklin Lakes, NJ, USA) using TruCountTM tubes (BD, Franklin Lakes, NJ, USA) in order to determine the absolute cell number of CellTrace+CD3+ cells in the samples as indicated in.52 Data were analyzed using FlowJo software (Treestar Inc.).
Statistics and bioinformatics analysis
t-SNE and clustering analysis were performed on flow cytometry data related to whole-blood immunophenotyping. Following doublet discrimination and debris exclusion based on light scattering characteristics, cell events of each sample were saved into Flow Cytometry Standard (FCS) files and imported into R/Bioconductor platform using the ‘flowCore’ package (https://www.bioconductor.org/packages/release/bioc/html/flowCore.html). Each matrix was down sampled to 5 × 103 events per sample to have a more efficient computation and then individually compensated and transformed using the Logicle transformation53. Next, for both P and NP patients, all the samples at each time point were concatenated in a single matrix. Fast interpolation-based t-SNE54 was used to visualize flow cytometry data into a low-dimensional space. FlowSOM metaclustering was performed on each matrix using the CATALYST package (https://github.com/HelenaLC/CATALYS) using default parameters. Cell type annotation was performed by looking at specific parameters (FSC-A, SSC-A, HLA-DR, CD45, CD3, CD4, CD14, CD16). Cell type quantities were reported as percentages of the total amount of cells using pie charts. Survival analyses were performed using ‘kmTCGA’ (https://rtcga.github.io/RTCGA/index.html) and ‘survminer’ (https://github.com/kassambara/survminer) packages. Logrank test was used to compare survival curves considering a p-value <.05 statistically significant. Figures were generated using the R packages ‘CATALYST’, ‘ggpubr’ (https://cran.r-project.org/web/packages/ggpubr/index.html), ‘flowCore’, and ‘ggplot2’ (https://ggplot2.tidyverse.org). The Mann-Whitney and Student’s t-test (paired or unpaired) were used to compare independently or not data obtained from P and NP patients. Pearson’s correlation coefficient was used to measure the statistical relationship between c-FLIP expression in circulating CD14+ cells and their immunosuppressive potential.
Results
Overall, 34 patients with advanced NSCLC who were treated with ICIs met the criteria for the final analysis. The clinical and biological characteristics are summarized in Table 1. According to RECIST evaluation [PD: progressive disease, SD: stable disease, PR: partial response], we classified patients as progressors (P) or non-progressors (NP) (Figure 1a).
Table 1.
Patients’ clinico-pathological characteristics at baseline.
| Characteristics | Patients N = 34%) |
NP N = 16%) |
P N = 18%) |
|---|---|---|---|
| Gender | |||
| Male | 23 (67.6) | 12 (75.0) | 11 (61.1) |
| Female | 11 (32.4) | 4 (25.0) | 7 (38.9) |
| Age years, median (range) | 72 (44–84) | 72.5 (58–82) | 72 (44–84) |
| ECOG performance status | |||
| 0 | 14 (41.2) | 8 (5.0) | 6 (33.3) |
| 1 | 17 (5.0) | 7 (43.8) | 10 (55.6) |
| 2 | 3 (8.8) | 1 (6.2) | 2 (11.1) |
| Smoker | |||
| Never | 7 (2.6) | 1 (6.2) | 6 (33.3) |
| Former | 20 (58.8) | 11 (68.8) | 9 (5.0) |
| Current | 7 (2.6) | 4 (25) | 3 (16.7) |
| Histology | |||
| Adenocarcinoma | 25 (73.5) | 11 (68.8) | 14 (77.8) |
| Squamous Carcinoma | 9 (26.5) | 5 (31.3) | 4 (22.2) |
| EGFR Status | |||
| Mutated | 1 (2.9) | 0 (.0) | 1 (5.6) |
| Wild Type | 33 (97.1) | 16 (1.0) | 17 (94.4) |
| PDL1% | |||
| <1% | 3 (8.8) | 1 (6.2) | 2 (11.1) |
| ≥1%-<50% | 15 (44.1) | 5 (31.3) | 10 (55.6) |
| ≥50% | 16 (47.1) | 10 (62.5) | 6 (33.3) |
| Immunotherapy Agent | |||
| Pembrolizumab | 16 (47.1) | 10 (62.5) | 6 (33.3) |
| Nivolumab | 14 (41.2) | 4 (25.0) | 10 (55.6) |
| Atezolizumab | 4 (11.7) | 2 (12.5) | 2 (11.1) |
| Line of immunotherapy | |||
| First | 16 (47.1) | 10 (62.5) | 6 (33.3) |
| Second | 18 (52.9) | 6 (37.5) | 12 (66.7) |
Abbreviations: N: number; NP: non progressor; P: Progressor; ECOG: Eastern Cooperative Oncology Group; EGFR: epidermal growth factor receptor; PD-L1: programmed death-ligand 1.
Figure 1.
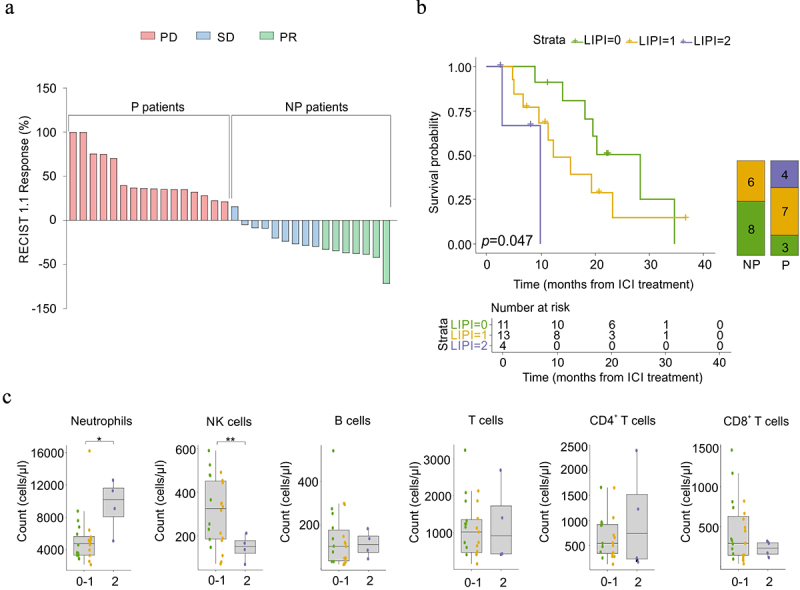
Clinical stratification of enrolled NSCLC patients and their immunological status before ICI immunotherapy.
(a) Best response to immunotherapy for target lesions by patient, based on the maximal percentage of tumor reduction, and the correspondent classification according to RECIST (Response Evaluation Criteria in Solid Tumors) 1.1. Two patients were not evaluable. PD: progressive disease, SD: stable disease, PR: partial response. (b) Kaplan – Meier curves (left) reporting the overall survival (OS, calculated from the date of start of ICI-treatment) of NSCLC patients (n = 28) stratified by lung immune prognostic index (LIPI) score at baseline as good (0, green), intermediate (1, yellow) and poor (2, purple). The overall log-rank test p = .0047. Bonferroni corrected LIPI 1 vs LIPI 0 p = 0.48, LIPI 1 vs LIPI 2 p = 0.72, LIPI 0 vs LIPI 2 p = .0147). Graph bar (right) reported NSCLC patient’s fractions with 0 (green), 1 (yellow), or 2 (purple) LIPI score in non-progressor (NP) and progressor (P) NSCLC patients (n = 28). The p-value (logrank test) was calculated to test differences among the three groups. (C) Box plots showing, at baseline, circulating neutrophils, NK cells, B cells, T cells, CD4+, and CD8+ T cells count in NSCLC patients (n = 28), classified by LIPI score (0 green, 1 yellow, 2 purple). Mann-Whitney or Student’s t-test. *p < .05, **p < .01.
ICI immunotherapy affects the blood immune landscape in non-progressor NSCLC patients
As expected, P patients had a significantly lower survival probability compared to NP patients (Figure 1a and Supplementary Figure S1a). Several studies have reported that the lung immune prognostic index (LIPI) score is one of the most promising and reliable tools for predicting ICI resistance in lung cancer patients.55 In agreement with these premises, NSCLC patients enrolled in this study with either “good” (0, green line) or “intermediate” (1, yellow line) LIPI score showed a significantly higher survival probability compared to patients identified with a “poor” LIPI score (2, purple line) at baseline (T0), regardless of whether they belonged to the P or NP group of patients (Figure 1b). All four patients showing the highest LIPI scores (purple line) were included in the P patient subgroup at T1, suggesting that they did not respond to ICI immunotherapy. Collectively, these results confirmed LIPI as a predictive score for ICI treatment in patients with NSCLC (Figure 1b).
The LIPI score considers the neutrophil/leukocytes-neutrophil (NLR) ratio and lactate dehydrogenase (LDH) plasma levels. We speculated that a deeper analysis of circulating immune soluble factors and cell subsets could provide additional candidates for predicting ICI efficacy in NSCLC patients. Indeed, neutrophils were significantly higher in patients showing a “poor” LIPI score (Figure 1c and Supplementary Figure S1b); among lymphocyte subsets, only natural killer (NK) cell count significantly diverged in this patient subgroup compared to patients characterized with either a “good” or an “intermediate” LIPI score (Figure 1c and Supplementary Figure S1b). In addition to the LIPI score, we also assessed other potential parameters, such as NLR, neutrophil-to-T lymphocytes (NTR), platelets-to-lymphocytes (PLR), neutrophils-to-CD4+ T cells, monocytes-to-lymphocytes (MLR), and monocyte-to-T lymphocyte (MTR) ratios; however, none of them at baseline were able to predict ICI response (Supplementary Figure S1c). Nevertheless, we detected a significant reduction in some of these parameters only in NP patients as a possible consequence of immunotherapy response, thus indicating the ability of PD-1/PD-L1 blockade not only to influence T cell effectiveness but also to alleviate systemic inflammation.
To investigate further the effects of ICI treatment, we quantified different pro-inflammatory cytokines in plasma samples before (T0) and after (T1) ICI immunotherapy. In the timeframe from T0 to T1, PD-1/PD-L1 inhibition significantly restrained the plasma levels of IL-6, IL-8, C–C chemokine ligand 4, IL-7, and IL-1β in NP patients, associating ICI responsiveness to a contraction in systemic inflammation (Figure 2). Other inflammation-associated cytokines, such as IL-8 and IL-1β, increased in P patients over time. Their plasma levels at T1 were higher than those detected in NP patients (Figure 2). Interestingly, IL-2 remained higher in NP patients at both observational time points (Figure 2). Collectively, only in NP patients the ICI-based immunotherapy promoted a time-dependent mitigation of several pro-inflammatory mediators whose basal levels were not predictive of response to immunotherapy.
Figure 2.
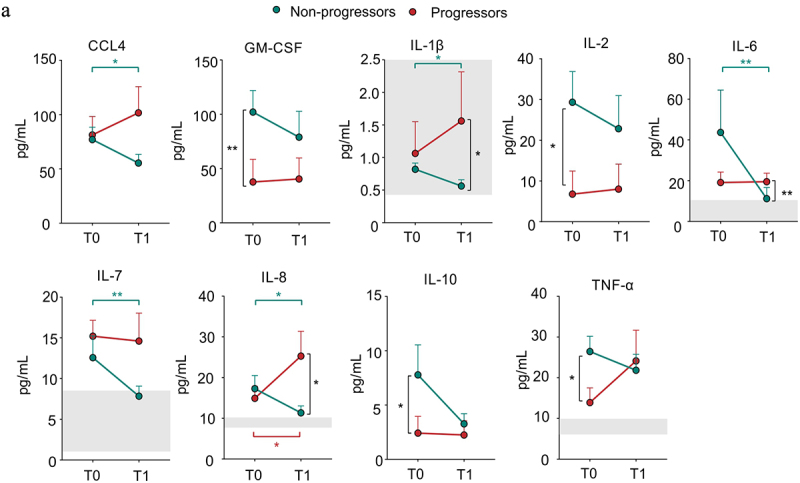
Effect of ICI immunotherapy on inflammation-associated cytokines in NSCLC patients.
(a) CCL4. GM-CSF, IL-1β, IL-2, IL-6, IL-7, IL-8, IL-10, TNF-α levels in plasma samples isolated from progressor (red) and non-progressor (green) NSCLC patients before (T0) and after (T1) ICI treatment (n = 34). The reference range of healthy subjects is reported with the light gray boxes. The data are shown as the mean ± SEM. Mann-Whitney and Wilcoxon test *p <.05, **p <.01. Stars and lines related to statistical analyses are indicated in: red for comparison between T0 and T1 in Progressor (P) patients; green for comparison between T0 and T1 in Non-Progressor (NP) patients; black for comparison between P and NP patients at either T0 (left) or T1 (right), respectively.
We then assessed the impact of ICI therapy on circulating immune cell populations. T-distributed stochastic neighbor embedding (t-SNE) and clustering analysis of peripheral blood revealed an increased proportion of T (from 13% to 16%), B (from 1% to 3%), and NK (from 4% to 5%) lymphocytes, with a concomitant decrease in neutrophils (from 68% to 62%) in NP patients after ICI treatment (Figure 3a upper panel and Supplementary Figure S2a). This unbiased analysis did not show any clear alteration of circulating populations in P patients, except for a weak increase in monocytes (from 9% to 10%) (Figure 3a lower panel and Supplementary Figure S2b). Through flow cytometry analysis, we confirmed an ICI-dependent increase in lymphocyte count in NP patients (Figure 3b). NK cells were higher in NP patients than in P patients, both before and after ICI treatment (Figure 3b). Similarly, CD8+CD4+ T cells increased only in the NP patient subgroup after ICI immunotherapy (Supplementary Figure S1b and Supplementary Figure S3a). Conversely, monocyte count significantly discriminated NP patients from P patients at the baseline (Figure 3b). To evaluate whether PD-1/PD-L1 inhibition could influence the monocyte compartment, we assessed the levels of three major monocyte populations: classic (CM, CD14+CD16− cells), non-classic (NCM, CD14dimCD16+ cells), and intermediate (IM, CD14+CD16+ cells) monocytes (Supplementary Figure S3b). Only the NCM count was significantly higher in NP patients than in P patients at T1, whereas, an increasing trend was detected for the other two monocytic subsets in NP patients at T0 (Supplementary Figure S3c). These data indicate that ICI therapy can reprogram systemic immunity by increasing lymphocyte frequency and altering myeloid cell composition.
Figure 3.
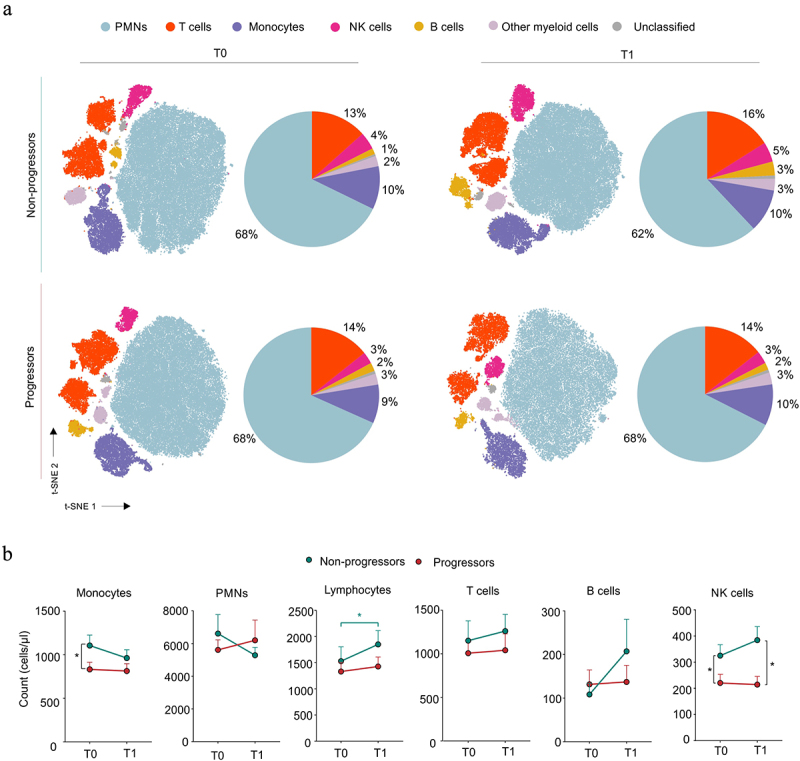
Effect of ICI immunotherapy on circulating immunological profile in NSCLC patients.
(a) t-distributed stochastic neighbor embedding (t-SNE) analysis on flow cytometry events following debris and doublet exclusion in non-progressor (NP) and progressor (P) patients before (T0) and after (T1) ICI treatment (T0-P and -NPn = 15, T1-P n = 9, T1-NPn = 15). Pie charts representing cell population proportions derived from clustering and t-SNE analysis for each group and time point. (b) Monocytes, PMNs, lymphocytes, T, B, and NK cell count in progressor (P, red) and non-progressor (NP, green) NSCLC patients before (T0) and after (T1) ICI treatment (n = 34). Mann–Whitney test. The data are shown as the mean ± SEM. *p < .05. Stars and lines related to statistical analyses are indicated in: red for comparison between T0 and T1 in Progressor (P) patients; green for comparison between T0 and T1 in Non-Progressor (NP) patients; black for comparison between P and NP patients at either T0 (left) or T1 (right), respectively.
ICI immunotherapy modifies immune-suppressive features of circulating M-MDSCs
One of the critical inducers of primary and secondary resistance to ICI therapy is the accumulation of several unconventional myeloid cell subsets with pro-tumoral functions defined by MDSCs.56 Therefore, we assessed the ability of PD-1/PD-L1 inhibition to affect the immunosuppressive functions of M-MDSCs identified as FLIP-overexpressing CD14+ cells. At baseline, NSCLC-derived CD14+ cells showed higher c-FLIP expression than healthy donors (HDs) (Figure 4a and Supplementary Figure S4a), which is in agreement with our previous report on PDAC patients.36 To further investigate FLIP in myeloid cells, we performed a t-SNE analysis of flow cytometry data from both P and NP patients, revealing a higher expression of c-FLIP in CD14+CD16−HLA-DRlow cells that resemble a conventional subgroup of M-MDSCs39 (Supplementary Figure S4b and Supplementary Figure S4c). We defined that c-FLIP mediates the acquisition of immunosuppressive features in monocytes.36 Accordingly, we observed a direct correlation between c-FLIP expression in CD14+ cells isolated from patients before ICI treatment and their ability to suppress the in vitro proliferation of activated CD3+ T lymphocytes (Figure 4b). FLIP expression in circulating M-MDSCs at T0 was similar in both P and NP patients, whereas after ICI treatment, a strong reduction in FLIP expression was detected only in the NP patient cohort (Figure 4c and Supplementary Figure S4a). However, this reduction did not involve specific conventional monocyte subsets (Supplementary Figure S4d). Furthermore, monocytes isolated from NP patients lost partially their ability to inhibit T-cell proliferation (Figure 4d). Notably, in vitro immunosuppressive activity was tested simultaneously by co-culturing thawed CD14+ cells isolated from patients at both T0 and T1 with the same in vitro activated allogeneic T cells, isolated from buffy coat, to standardize functional evaluation. Collectively, our data highlight that ICI therapy affects M-MDSC-dependent immunosuppressive activity probably due to the decrease of c-FLIP expression.
Figure 4.
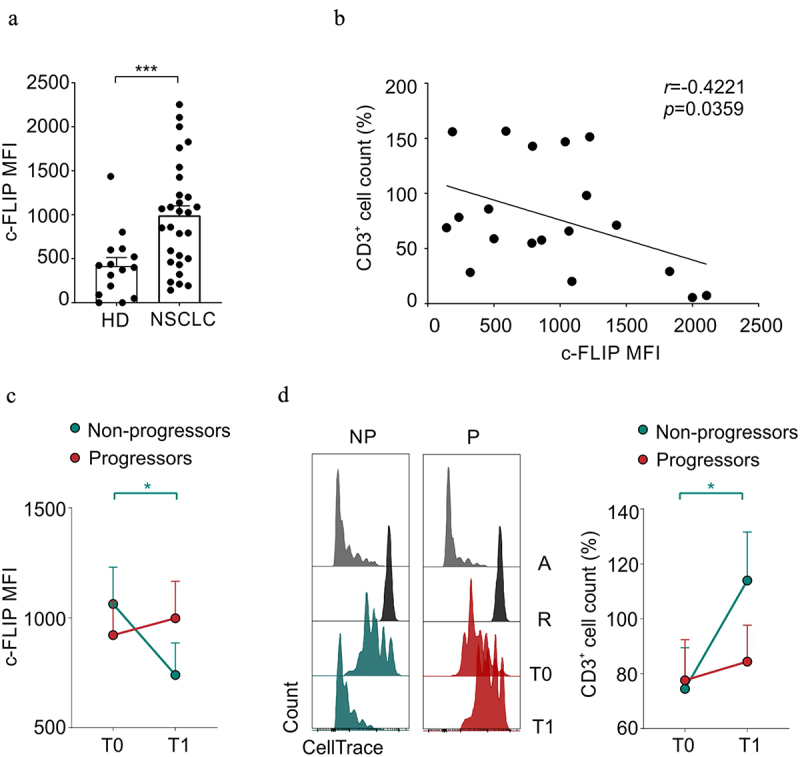
Effect of ICI immunotherapy on immunosuppressive features of M-MDSCs in NSCLC patients.
(a) c-FLIP expression evaluated by FACS analysis and reported as mean fluorescence intensity (MFI), FMO (fluorescence minus one) corrected, in circulating CD14+ cells from healthy donors (HD) and NSCLC patients. Mann–Whitney test. (b) Circulating CD14+ cells isolated from NSCLC patients before ICI treatment were co-cultured with HD-derived, α-CD3 and α-CD28 activated PBMCs in order to evaluate their immunosuppressive ability. The percentage of CellTrace+CD3+ T lymphocytes cell count deriving from immunosuppression assays was correlated with CD14+ c-FLIP expression evaluated as FMO corrected MFI. Pearson r correlation. (c) FMO corrected c-FLIP MFI of CD14+ cells in progressor (P, green) and non-progressor (NP, red) patients before (T0) and after (T1) ICI treatment (T0-P n = 16, T1-P n = 10, T0-NPn = 15, T1-NPn = 14). Mann-Whitney and Wilcoxon test. (d) Representative proliferation peaks of CellTrace+CD3+ T lymphocytes following the co-culture with circulating CD14+ cells isolated from progressor (red) and non-progressor (green) NSCLC patients before (T0) and after (T1) ICI treatment (left) (A: Activated control, R: resting control). Percentage of CellTrace+CD3+ T lymphocytes cell count following the co-culture with circulating CD14+ cells isolated from progressor (red) and non-progressor (green) NSCLC patients before (T0) and after (T1) ICI treatment (T0-P n = 8, T1-P n = 9, T0-NPn = 12, T1-NPn = 12) (right). All values are normalized on activated T cells in the absence of myeloid cells. Mann-Whitney and Wilcoxon test. The data are shown as the mean ± SEM. *p < .05, ****p < .0001. Stars and lines related to statistical analyses are indicated in green or red if they referred to non-progressor or progressor patients, respectively.
Discussion
MDSCs share morphological features and common lineage markers with conventional monocytes and granulocytes; however, they have tolerogenic and pro-tumoral properties.24,56,57 Since phenotypical analysis is not a conclusive to identify MDSCs, a position manuscript of the research community defined the evaluation of suppressive activity and the identification of critical transcription factors and regulators as mandatory aspects that must be tested to support the MDSC definition.22 Following these recommendations, in 2019, our group published a case report of an NSCLC patient treated with durvalumab as maintenance therapy after chemotherapy and radiotherapy, which showed a decrease in MDSCs after two administrations of anti-PD-L1 antibody. The therapeutic impact of durvalumab was thus associated with precise functional impairment of MDSCs.58 Howeverto date, there are limited and controversial preclinical and clinical data regarding the effect of ICI on MDSC frequency and function.32
To our knowledge, this is the first prospective immune monitoring study to evaluate the effects of ICI therapy on modulating MDSC-associated immunosuppression and inflammation. Our findings indicate that immunotherapy treatment leads to a contraction of circulating factors such as CCL4, IL-1β, IL-6, IL-7, and IL-8 in the NP patient cohort exclusively. All these soluble factors were identified as potential targets to improve immunotherapy effectiveness,59–62 highlighting how the repression of these pro-inflammatory cytokines is essential to trigger a successful anti-tumor immune response. In particular, IL-8 appears to play a critical role on defining ICI effectiveness in our patient cohort since in the time window between T0 and T1 it was significantly decreased in plasma of NP patients while significantly deepening in P patients. On the other hand, high levels of GM-CSF, IL-2, IL-10 and TNFα, before ICI treatment were able to define NP patients and, therefore, they may be served as potential predictive biomarkers for response to immunotherapy. Certainly, it is mandatory to validate all these parameters using independent and large patient cohorts, as well as testing these parameters in different cancers. Despite the limited number of patients tested in this work, our findings revealed that P patients never showed ICI-dependent alterations in tested circulating factors, except for IL-8, suggesting that, probably, these patients are immune unresponsive. A deeper analysis on the cell source of these inflammation-associated mediators is worthy of future studies with the aim to develop both innovative therapeutic approaches and more efficient tools for early diagnosis of resistance to therapy.
We previously reported that c-FLIP-expressing myeloid cells released several inflammation-associated molecules such IL-6 and TNF-α by a “steered” NF-κB activation, which also resulted in enhanced STAT3-signaling activation.36,37 Furthermore, c-FLIP promotes the acquisition of immunosuppression-associated features that can explain the inhibition of T cell activation and proliferation, i.e., receptor–ligand interactions (i.e., PD-L1 axis), metabolic pathways (i.e., IDO1-dependent tryptophan metabolism pathway), and production of cytokines (i.e., IL-6).36 Thus, we exploited c-FLIP detection to monitor changes in MDSCs by immunotherapy. In line with our previous findings in the context of both PDAC36 and COVID-1937 patients, we found high level of c-FLIP expression in NSCLC patient-derived monocytes before immunotherapy treatment. Moreover, the immunosuppressive functions of monocytes isolated from patients with NSCLC directly correlated with the expression of c-FLIP. It is important to highlight that c-FLIP expression by t-SNE analysis was preferentially restricted to a classical monocyte subset characterized as CD14+HLA-DRlow cells, which identify a specific MDSC subgroup (also defined as MDSC4), which can discriminate PDAC patients with metastatic disease.39 ICI treatment potentially reduced c-FLIP expression in circulating monocytes and also their inhibitory properties only in NP patients. Coupled with FLIP expression of this pro-tumor cell subset, NP patients showed a significant increase in circulating lymphocytes, especially NK cells and CD8+CD4+ T cells. Recently, an enrichment in CD4+CD8+ T cells among patients who ultimately responded to ICI has been reported.63 Notably, double-positive T cells were characterized by potent tumor major histocompatibility complex (MHC)-dependent reactivity, suggesting a critical role of these cells in specific ICI-induced effector lymphocytes.63
The clinical accomplishment of cancer immunotherapy has paved the way for a more comprehensive view of cancer, highlighting the critical role of immune cells with immunosuppressive functions such as MDSCs. T cell functions can be intrinsically affected by the engagement of inhibitory receptors, as well as by nutrient and growth factor competition with several cell subsets in the TME, including immunosuppressive myeloid cells. Therefore, targeting these immune inhibitory cell subsets is a potential therapeutic strategy to enhance cancer immunotherapy. For instance, targeting the macrophage scavenger receptor MARCO or its induction through blockade of IL37 receptor has been demonstrated to be an efficient approach to reprogram suppressive TME-infiltrating macrophages in NSCLC immune landscape and to restore T-cell and NK-cell antitumor activities.64 Other potential therapeutic strategies to limit immunosuppressive myeloid cells are metabolic pathways such as kynurenines derived from the IDO1-associated pathway,65,66 degradation of arginine, and generation of RNS,67,68 components of the adenosine pathway,69 and glutamine metabolism pathway.70 Interestingly, some of these targets, such as IDO1 and ARG1, can also be targeted by both active71,72 and passive73 immunotherapies. Finally, aiming at inflammatory mediators, such as IL-6, might be an effective strategy to enforce and boost anti-tumor, ICI-induced immunity by decreasing MDSCs and macrophages within the tumor mass.60 However, a complete deactivation or elimination of myeloid cells is still a challenging approach because of the inherent plasticity of these elements, which can alter both their phenotype and functions during cancer progression, and, overall, are influenced in an organ-specific manner. Nonetheless, this is viewed by many as the next step in cancer immunotherapy development.
Despite reproducible positive results, only a minority (<20%) of the patients show long-term benefit from ICI immunotherapy and most will progress at any time during treatment; furthermore, a non-negligible proportion of patients receiving ICI do not respond to treatment despite high PD-L1 expression. Current biomarkers for predicting the response to ICI therapy are usually based on biopsies taken from solid tumors, including the PD-L1 status. These biomarkers require invasive techniques that pose significant challenges that can be overcome using liquid biopsies to analyze circulating immune parameters. Our study suggests new potential and more advantageous targets, including monocyte and NK count, as well as inflammation-associated soluble mediators such as TNF-α, GM-CSF, and IL-2. Furthermore, we described a rapid variation of additional immunological features following immunotherapy only in NP patients, including c-FLIP expression in monocytes, CD4+CD8+ T and NK lymphocyte counts, and inflammatory cytokines, such as IL-6. Recently, blood-based proteomic screening revealed a predictive signature able to stratify NP and P patients, in which IL-6, neutrophil-related proteins, janus kinase-STAT, and NF-κB signaling pathways were identified as key players in promoting resistance to ICI.74 Our study had several limitations. First, the enrolled patient cohort was small for biomarker discovery and required validation in an independent cohort. Second, the cohort included patients who were treated with different ICIs. Third, flow cytometry analysis was based on a limited number of phenotypic markers, precluding the identification of T cell functional stage and the enumeration of other leukocyte subsets, such as dendritic cells, T regulatory cells, and T helper subsets, as well as the detection of c-FLIP in other circulating leukocytes. Fourth, alterations in the gene signature of c-FLIP-overexpressing cells induced by ICI were not evaluated.
In conclusion, our study indicates that the clinical impact of ICI promotes a modulation of the immune landscape by limiting systemic inflammation and controlling MDSC-dependent immunosuppression.
Supplementary Material
Acknowledgments
The authors thank the patients who contributed samples to the study and research nurses for collecting of patients’ samples. We thank Roza Maria Barouni, Cristina Anselmi, Giulio Fracasso, Tiziana Cestari, Eliana Urbini, Amalia Montini, Nicolas Binetti, Giammarco Ciresola, Morena Martini, Fiorenza Paiola, Elena Lucchini, Claudia Pizzoli, Elena Chiesa, Oretta Gabrielli, Nadia Brutti, Monica Brentegani, and Raffaella Quitadamo for their excellent technical work at Immunology Section. We also thank Marta Donini of General Pathology Section for her outstanding work.
Funding Statement
This work was jointly supported by both Stefano Ugel grants of the Fondazione Italiana per la Ricerca sul Cancro (AIRC) (MFAG project: 21509), PRIN program of Italian Ministry of University and Research (MUR, CUP: B38D19000140006), Fondazione Cariverona (ENACT project) and PNRR programs of the Italian MUR (Project “National Center for Gene Therapy and Drugs based on RNA Technology”, application code CN00000041, Mission 4, Component 2 Investment 1.4, funded from the European Union – NextGeneration EU, MUR Directorial Decree No. 1035 of 17 June 2022, CUP B33C22000630001); Vincenzo Bronte grants funded by Fondazione Cariverona (project call, 2017), Fondazione AIRC (IG project: 23788), Cancer Research Institute (Clinic and Laboratory Integration Program, CLIP 2020), and PRIN program of Italian MUR (CUP: B38D19000260006). Annalisa Adamo was supported by AIRC/FIRC fellowship call 2019; Chiara Musiu was supported by AIRC fellowship call 2022. Emilio Bria. is supported by Fondazione AIRC (IG project: 20583) and by Institutional funds of Università Cattolica del Sacro Cuore (UCSC-project D1). Carmine Carbone is supported by Fondazione AIRC (MFAG project: 23681). Francesco De Sanctis is supported by the PRIN program of Italian MUR (CUP: B39J22001200001). Fang Qi is sponsored by the China scholarship Council (No. 202108520059) at the University of Verona.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Authors’ Contributions
A.A., C.F., S.P., V.B., and S.U. conceptualized the study; C.F., A.A., S.P., and S.U. designed the methodology; C.F., A.A, O.O., D.R., A.V., S.C., C.C., G.P., F.P., Si.D., C.M., Y.H., T.W., and S.U. performed experiments; S.C. and L.G. conducted computational analysis and interpreted data; S.P., L.B., G.S., I.J., M.S., U.C.I., E.B., and M.M were responsible for biological specimens and patients’ data collection; C.C. and G.P. performed cytokine detection; A.A., S.P., F.Q., D.W., S.S., and F.D.S. interpreted data, A.A., S.C., P.S., St.D. M.A.C., E.B., and M.M co-designed the study, interpreted the data, and contributed to writing the paper; V.B. and S.U. directed the project, designed the study, interpreted data, supervised research, wrote the manuscript, and acquired the funding. All authors read, revised, and approved the final manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study as well as reagents and samples are available from the senior authors on reasonable request and under material transfer agreement.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2023.2253644
References
- 1.Cancer Genome Atlas Research N . Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–12. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh T, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang B, Liu Z, Jin L, Xing R, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24(7):978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 5.Lavin Y, Kobayashi S, Leader A, Amir EAD, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169(4):750–765.e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larroquette M, Guegan JP, Besse B, Cousin S, Brunet M, Le Moulec S, Le Loarer F, Rey C, Soria J-C, Barlesi F, et al. Spatial transcriptomics of macrophage infiltration in non-small cell lung cancer reveals determinants of sensitivity and resistance to anti-PD1/PD-L1 antibodies. J Immunother Cancer. 2022;10(5):e003890. doi: 10.1136/jitc-2021-003890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper AJ, Sequist LV, Lin JJ.. Third-generation EGFR and ALK inhibitors: mechanisms of resistance and management. Nat Rev Clin Oncol. 2022;19(8):499–514. doi: 10.1038/s41571-022-00639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang CY, Yang JC, Yang PC. Precision management of advanced non–small cell lung cancer. Annu Rev Med. 2020;71(1):117–136. doi: 10.1146/annurev-med-051718-013524. [DOI] [PubMed] [Google Scholar]
- 9.Yuan M, Huang LL, Chen JH, Wu J, Xu Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduction Targeted Ther. 2019;4(1):61. doi: 10.1038/s41392-019-0099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye L, Creaney J, Redwood A, Robinson B. The Current lung cancer neoantigen landscape and implications for therapy. J Thorac Oncol. 2021;16(6):922–932. doi: 10.1016/j.jtho.2021.01.1624. [DOI] [PubMed] [Google Scholar]
- 11.Grant MJ, Herbst RS, Goldberg SB. Selecting the optimal immunotherapy regimen in driver-negative metastatic NSCLC. Nat Rev Clin Oncol. 2021;18(10):625–644. doi: 10.1038/s41571-021-00520-1. [DOI] [PubMed] [Google Scholar]
- 12.Chaft JE, Rimner A, Weder W, Azzoli CG, Kris MG, Cascone T. Evolution of systemic therapy for stages I–III non-metastatic non-small-cell lung cancer. Nat Rev Clin Oncol. 2021;18(9):547–557. doi: 10.1038/s41571-021-00501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang AC, Herbst RS. Frontline immunotherapy for NSCLC - the tale of the tail. Nat Rev Clin Oncol. 2020;17(2):73–74. doi: 10.1038/s41571-019-0317-y. [DOI] [PubMed] [Google Scholar]
- 14.Iams WT, Porter J, Horn L. Immunotherapeutic approaches for small-cell lung cancer. Nat Rev Clin Oncol. 2020;17(5):300–312. doi: 10.1038/s41571-019-0316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacquelot N, Yamazaki T, Roberti MP, Duong CPM, Andrews MC, Verlingue L, Ferrere G, Becharef S, Vétizou M, Daillère R, et al. Sustained type I interferon signaling as a mechanism of resistance to PD-1 blockade. Cell Res. 2019;29(10):846–861. doi: 10.1038/s41422-019-0224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27(4):462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 21.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19(2):108–119. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7(1):12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ugel S, De Sanctis F, Mandruzzato S, Bronte V. Tumor-induced myeloid deviation: when myeloid-derived suppressor cells meet tumor-associated macrophages. J Clin Invest. 2015;125(9):3365–3376. doi: 10.1172/JCI80006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Sanctis F, Solito S, Ugel S, Molon B, Bronte V, Marigo I. Mdscs in cancer: Conceiving new prognostic and therapeutic targets. Biochim Biophys Acta. 2016;1865(1):35–48. doi: 10.1016/j.bbcan.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Groth C, Hu X, Weber R, Fleming V, Altevogt P, Utikal J, Umansky V. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br J Cancer. 2019;120(1):16–25. doi: 10.1038/s41416-018-0333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5(8):641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 28.Hofer F, Di Sario G, Musiu C, Sartoris S, De Sanctis F, Ugel S. A complex metabolic network confers immunosuppressive functions to myeloid-derived suppressor cells (MDSCs) within the tumour microenvironment. Cells. 2021;10(10):2700. doi: 10.3390/cells10102700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. 2021;21(8):485–498. doi: 10.1038/s41577-020-00490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trovato R, Cane S, Petrova V, Sartoris S, Ugel S, De Sanctis F. The engagement between MDSCs and metastases: Partners in crime. Front Oncol. 2020;10:165. doi: 10.3389/fonc.2020.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solito S, Marigo I, Pinton L, Damuzzo V, Mandruzzato S, Bronte V. Myeloid-derived suppressor cell heterogeneity in human cancers. Ann NY Acad Sci. 2014;1319(1):47–65. doi: 10.1111/nyas.12469. [DOI] [PubMed] [Google Scholar]
- 32.Peranzoni E, Ingangi V, Masetto E, Pinton L, Marigo I. Myeloid cells as clinical biomarkers for immune checkpoint blockade. Front Immunol. 2020;11:1590. doi: 10.3389/fimmu.2020.01590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber R, Fleming V, Hu X, Nagibin V, Groth C, Altevogt P, Utikal J, Umansky V. Myeloid-derived suppressor cells hinder the anti-cancer activity of immune checkpoint inhibitors. Front Immunol. 2018;9:1310. doi: 10.3389/fimmu.2018.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grover A, Sanseviero E, Timosenko E, Gabrilovich DI. Myeloid-derived suppressor cells: A propitious road to clinic. Cancer Discov. 2021;11(11):2693–2706. doi: 10.1158/2159-8290.CD-21-0764. [DOI] [PubMed] [Google Scholar]
- 35.Haverkamp JM, Smith AM, Weinlich R, Dillon C, Qualls J, Neale G, Koss B, Kim Y, Bronte V, Herold M, et al. Myeloid-derived suppressor activity is mediated by monocytic lineages maintained by continuous inhibition of extrinsic and intrinsic death pathways. Immunity. 2014;41(6):947–959. doi: 10.1016/j.immuni.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiore A, Ugel S, De Sanctis F, Sandri S, Fracasso G, Trovato R, Sartoris S, Solito S, Mandruzzato S, Vascotto F, et al. Induction of immunosuppressive functions and NF-κB by FLIP in monocytes. Nat Commun. 2018;9(1):5193. doi: 10.1038/s41467-018-07654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musiu C, Caligola S, Fiore A, Lamolinara A, Frusteri C, Del Pizzo FD, De Sanctis F, Canè S, Adamo A, Hofer F, et al. Fatal cytokine release syndrome by an aberrant FLIP/STAT3 axis. Cell Death Differ. 2022;29(2):420–438. doi: 10.1038/s41418-021-00866-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasquez-Dunddel D, Pan F, Zeng Q, Gorbounov M, Albesiano E, Fu J, Blosser RL, Tam AJ, Bruno T, Zhang H, et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J Clin Invest. 2013;123(4):1580–1589. doi: 10.1172/JCI60083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trovato R, Fiore A, Sartori S, Canè S, Giugno R, Cascione L, Paiella S, Salvia R, De Sanctis F, Poffe O, et al. Immunosuppression by monocytic myeloid-derived suppressor cells in patients with pancreatic ductal carcinoma is orchestrated by STAT3. J Immunother Cancer. 2019;7(1):255. doi: 10.1186/s40425-019-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kluger HM, Tawbi HA, Ascierto ML, Bowden M, Callahan MK, Cha E, Chen HX, Drake CG, Feltquate DM, Ferris RL, et al. Defining tumor resistance to PD-1 pathway blockade: recommendations from the first meeting of the SITC immunotherapy resistance taskforce. J Immunother Cancer. 2020;8(1):e000398. doi: 10.1136/jitc-2019-000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoenfeld AJ, Antonia SJ, Awad MM, Felip E, Gainor J, Gettinger SN, Hodi FS, Johnson ML, Leighl NB, Lovly CM, et al. Clinical definition of acquired resistance to immunotherapy in patients with metastatic non-small-cell lung cancer. Ann Oncol. 2021;32(12):1597–1607. doi: 10.1016/j.annonc.2021.08.2151. [DOI] [PubMed] [Google Scholar]
- 42.Fayad L, Keating MJ, Reuben JM, O’Brien S, Lee B-N, Lerner S, Kurzrock R. Interleukin-6 and interleukin-10 levels in chronic lymphocytic leukemia: correlation with phenotypic characteristics and outcome. Blood. 2001;97(1):256–263. doi: 10.1182/blood.V97.1.256. [DOI] [PubMed] [Google Scholar]
- 43.Bronte V, Ugel S, Tinazzi E, Vella A, De Sanctis F, Canè S, Batani V, Trovato R, Fiore A, Petrova V, et al. Baricitinib restrains the immune dysregulation in patients with severe COVID-19. J Clin Invest. 2020;130(12):6409–6416. doi: 10.1172/JCI141772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pradhan AD, Manson JE, Rifai N, Buring JE, Ridker PM.. C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus. JAMA. 2001;286(3):327–334. doi: 10.1001/jama.286.3.327. [DOI] [PubMed] [Google Scholar]
- 45.Silva EM, Mariano VS, Pastrez PRA, Pinto MC, Castro AG, Syrjanen KJ, Longatto-Filho A. High systemic IL-6 is associated with worse prognosis in patients with non-small cell lung cancer. PLoS One. 2017;12(7):e0181125. doi: 10.1371/journal.pone.0181125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Neill CM, Lu C, Corbin KL, Sharma PR, Dula SB, Carter JD, Ramadan JW, Xin W, Lee JK, Nunemaker CS, et al. Circulating levels of IL-1B+IL-6 cause ER stress and dysfunction in islets from prediabetic male mice. Endocrinology. 2013;154(9):3077–3088. doi: 10.1210/en.2012-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohashi A, Uemura Y, Yoshimori M, Wada N, Imadome K-I, Yudo K, Koyama T, Shimizu N, Nishio M, Arai A, et al. The plasma level of interleukin-1β can be a biomarker of angiopathy in systemic chronic active Epstein–Barr virus infection. Front Microbiol. 2022;13:874998. doi: 10.3389/fmicb.2022.874998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrajoli A, Keating MJ, Manshouri T, Giles FJ, Dey A, Estrov Z, Koller CA, Kurzrock R, Thomas DA, Faderl S, et al. The clinical significance of tumor necrosis factor-α plasma level in patients having chronic lymphocytic leukemia. Blood. 2002;100(4):1215–1219. doi: 10.1182/blood.V100.4.1215.h81602001215_1215_1219. [DOI] [PubMed] [Google Scholar]
- 49.Lundstrom W, Fewkes NM, Mackall CL. IL-7 in human health and disease. Semin Immunol. 2012;24(3):218–224. doi: 10.1016/j.smim.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang P, Liu D, Li L, Guo X, Ye Y, Li Y, Jiang Q, Lin S, Yuan Q. Interleukin 8 in plasma is an efficacy marker for advanced non-small cell lung cancer treated with hypofractionated radiotherapy and PD-1 blockade. Cytokine. 2023;163:156133. doi: 10.1016/j.cyto.2023.156133. [DOI] [PubMed] [Google Scholar]
- 51.Borilova Linhartova P, Kavrikova D, Tomandlova M, Poskerova H, Rehka V, Dušek L, Izakovicova Holla L. Differences in interleukin-8 plasma levels between diabetic patients and healthy individuals independently on their periodontal status. Int J Mol Sci. 2018;19(10):3214. doi: 10.3390/ijms19103214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solito S, Pinton L, De Sanctis F, Ugel S, Bronte V, Mandruzzato S, Marigo I. Methods to measure MDSC immune suppressive activity in vitro and in vivo. Curr Protoc Immunol. 2019;124(1):e61. doi: 10.1002/cpim.61. [DOI] [PubMed] [Google Scholar]
- 53.Parks DR, Roederer M, Moore WA. A new “Logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A. 2006;69(6):541–551. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- 54.Linderman GC, Rachh M, Hoskins JG, Steinerberger S, Kluger Y. Fast interpolation-based t-SNE for improved visualization of single-cell RNA-seq data. Nat Methods. 2019;16(3):243–245. doi: 10.1038/s41592-018-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mezquita L, Auclin E, Ferrara R, Charrier M, Remon J, Planchard D, Ponce S, Ares LP, Leroy L, Audigier-Valette C, et al. Association of the lung immune prognostic index with immune checkpoint inhibitor outcomes in patients with advanced non–small cell lung cancer. JAMA Oncol. 2018;4(3):351–357. doi: 10.1001/jamaoncol.2017.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long H, Jia Q, Wang L, Fang W, Wang Z, Jiang T, Zhou F, Jin Z, Huang J, Zhou L, et al. Tumor-induced erythroid precursor-differentiated myeloid cells mediate immunosuppression and curtail anti-PD-1/PD-L1 treatment efficacy. Cancer Cell. 2022;40(6):674–693.e7. doi: 10.1016/j.ccell.2022.04.018. [DOI] [PubMed] [Google Scholar]
- 57.Li R, Salehi-Rad R, Crosson W, Momcilovic M, Lim RJ, Ong SL, Huang ZL, Zhang T, Abascal J, Dumitras C, et al. Inhibition of granulocytic myeloid-derived suppressor cells overcomes resistance to immune checkpoint inhibition in LKB1-deficient non–small cell lung cancer. Cancer Res. 2021;81(12):3295–3308. doi: 10.1158/0008-5472.CAN-20-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bertelli G, Trovato R, Ugel S, Bria E, Milella M, Bronte V, Pilotto S. Characterization of myeloid-derived suppressor cells in a patient with lung adenocarcinoma Undergoing durvalumab treatment: A case report. Clin Lung Cancer. 2019;20(4):e514–e516. doi: 10.1016/j.cllc.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 59.Liu H, Zhao Q, Tan L, Wu X, Huang R, Zuo Y, Chen L, Yang J, Zhang Z-X, Ruan W, et al. Neutralizing IL-8 potentiates immune checkpoint blockade efficacy for glioma. Cancer Cell. 2023;41(4):693–710.e8. doi: 10.1016/j.ccell.2023.03.004. [DOI] [PubMed] [Google Scholar]
- 60.Hailemichael Y, Johnson DH, Abdel-Wahab N, Foo WC, Bentebibel S-E, Daher M, Haymaker C, Wani K, Saberian C, Ogata D, et al. Interleukin-6 blockade abrogates immunotherapy toxicity and promotes tumor immunity. Cancer Cell. 2022;40(5):509–523.e6. doi: 10.1016/j.ccell.2022.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L, Liu YD, Zhan YT, Zhu Y-H, Li Y, Xie D, Guan X-Y. High levels of CCL2 or CCL4 in the tumor microenvironment predict unfavorable survival in lung adenocarcinoma. Thorac Cancer. 2018;9(7):775–784. doi: 10.1111/1759-7714.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaplanov I, Carmi Y, Kornetsky R, Shemesh A, Shurin GV, Shurin MR, Dinarello CA, Voronov E, Apte RN. Blocking IL-1β reverses the immunosuppression in mouse breast cancer and synergizes with anti–PD-1 for tumor abrogation. Proc Natl Acad Sci USA. 2019;116(4):1361–1369. doi: 10.1073/pnas.1812266115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schad SE, Chow A, Mangarin L, Pan H, Zhang J, Ceglia N, Caushi JX, Malandro N, Zappasodi R, Gigoux M, et al. Tumor-induced double positive T cells display distinct lineage commitment mechanisms and functions. J Exp Med. 2022;219(6):219 (6. doi: 10.1084/jem.20212169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.La Fleur L, Botling J, He F, Pelicano C, Zhou C, He C, Palano G, Mezheyeuski A, Micke P, Ravetch JV, et al. Targeting MARCO and IL37R on immunosuppressive macrophages in lung cancer blocks regulatory T cells and supports cytotoxic lymphocyte function. Cancer Res. 2021;81(4):956–967. doi: 10.1158/0008-5472.CAN-20-1885. [DOI] [PubMed] [Google Scholar]
- 65.Campesato LF, Budhu S, Tchaicha J, Weng C-H, Gigoux M, Cohen IJ, Redmond D, Mangarin L, Pourpe S, Liu C, et al. Blockade of the AHR restricts a Treg-macrophage suppressive axis induced by L-Kynurenine. Nat Commun. 2020;11(1):4011. doi: 10.1038/s41467-020-17750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Serafini M, Torre E, Aprile S, Grosso ED, Gesù A, Griglio A, Colombo G, Travelli C, Paiella S, Adamo A, et al. Discovery of highly potent benzimidazole derivatives as Indoleamine 2,3-Dioxygenase-1 (IDO1) inhibitors: From structure-based virtual screening to in vivo pharmacodynamic activity. J Med Chem. 2020;63(6):3047–3065. doi: 10.1021/acs.jmedchem.9b01809. [DOI] [PubMed] [Google Scholar]
- 67.Molon B, Ugel S, Del Pozzo F, Soldani C, Zilio S, Avella D, De Palma A, Mauri P, Monegal A, Rescigno M, et al. Chemokine nitration prevents intratumoral infiltration of antigen-specific T cells. J Exp Med. 2011;208(10):1949–1962. doi: 10.1084/jem.20101956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Sanctis F, Lamolinara A, Boschi F, Musiu C, Caligola S, Trovato R, Fiore A, Frusteri C, Anselmi C, Poffe O, et al. Interrupting the nitrosative stress fuels tumor-specific cytotoxic T lymphocytes in pancreatic cancer. J Immunother Cancer. 2022;10(1):e003549. doi: 10.1136/jitc-2021-003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vigano S, Alatzoglou D, Irving M, Ménétrier-Caux C, Caux C, Romero P, Coukos G. Targeting Adenosine in cancer immunotherapy to Enhance T-Cell function. Front Immunol. 2019;10:925. doi: 10.3389/fimmu.2019.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oh MH, Sun IH, Zhao L, Leone RD, Sun I-M, Xu W, Collins SL, Tam AJ, Blosser RL, Patel CH, et al. Targeting glutamine metabolism enhances tumor-specific immunity by modulating suppressive myeloid cells. J Clin Invest. 2020;130(7):3865–3884. doi: 10.1172/JCI131859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aaboe Jorgensen M, Ugel S, Linder Hubbe M, Carretta M, Perez-Penco M, Weis-Banke SE, Martinenaite E, Kopp K, Chapellier M, Adamo A, et al. Arginase 1–based immune modulatory vaccines induce anticancer immunity and synergize with anti–PD-1 checkpoint blockade. Cancer Immunol Res. 2021;9(11):1316–1326. doi: 10.1158/2326-6066.CIR-21-0280. [DOI] [PubMed] [Google Scholar]
- 72.Kjeldsen JW, Lorentzen CL, Martinenaite E, Ellebaek E, Donia M, Holmstroem RB, Klausen TW, Madsen CO, Ahmed SM, Weis-Banke SE, et al. A phase 1/2 trial of an immune-modulatory vaccine against IDO/PD-L1 in combination with nivolumab in metastatic melanoma. Nat Med. 2021;27(12):2212–2223. doi: 10.1038/s41591-021-01544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cane S, Barouni RM, Fabbi M, Cuozzo J, Fracasso G, Adamo A, Ugel S, Trovato R, De Sanctis F, Giacca M, et al. Neutralization of NET-associated human ARG1 enhances cancer immunotherapy. Sci Transl Med. 2023;15(687):eabq6221. doi: 10.1126/scitranslmed.abq6221. [DOI] [PubMed] [Google Scholar]
- 74.Harel M, Lahav C, Jacob E, Dahan N, Sela I, Elon Y, Raveh Shoval S, Yahalom G, Kamer I, Zer A, et al. Longitudinal plasma proteomic profiling of patients with non-small cell lung cancer undergoing immune checkpoint blockade. J Immunother Cancer. 2022;10(6):e004582. doi: 10.1136/jitc-2022-004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study as well as reagents and samples are available from the senior authors on reasonable request and under material transfer agreement.


