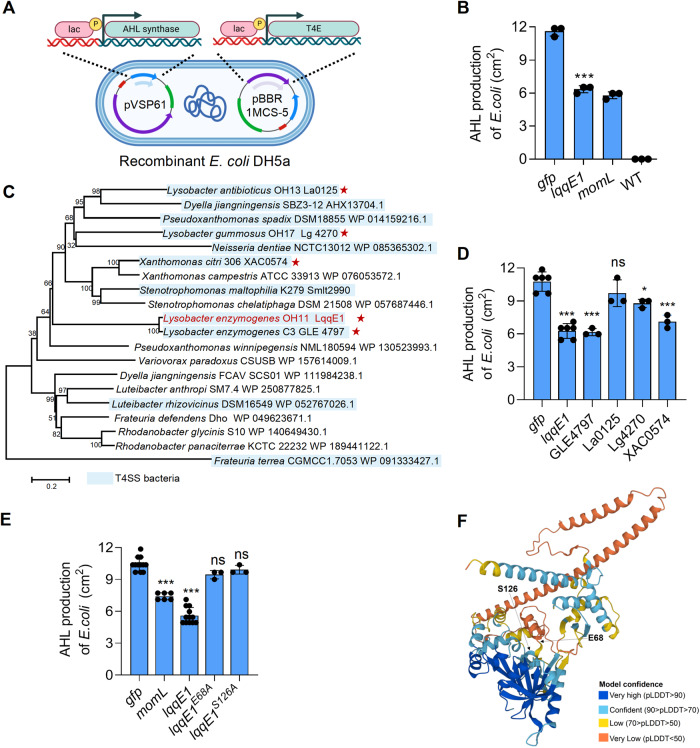
Fig. 3. Identification of key amino residues responsible for LqqE1 functioning as a quorum-quenching protein.
A A schematic map showing how to co-express the lqqE1 gene with AHL synthase genes by plasmids in E. coli. B Co-expression of the lqqE1 gene with the AHL synthase PcoI gene of P. fluorescens 2P24 in E. coli significantly reduced AHL production. Both the wild-type E. coli and the plasmid harboring gfp were used as negative controls; MomL, a known AHL-degrading enzyme, served as a positive control. C The distribution of the LqqE1 in diverse bacterial genomes. The gene name/NCBI accession number of the LqqE1 homologs was listed after the bacterial species. T4ASS-producing bacteria were highlighted by light blue. Red asterisk indicated these LqqE1 homologs were selected for testing their AHL-quenching activity. D Three selected LqqE1 homologs (GLE4749 from L. enzymogenes C3, Lg4270 from L. gummosus OH17, and XAC0574 from Xanthomonas citri 306) showed an AHL-quenching activity when their genes were co-expressed with the PcoI gene in E. coli, according to the protocol shown in (A). E Co-expression of the pcoI gene with lqqE1 variant genes in E. coli showed that two residues (E68, S126) are required for LqqE1 to execute AHL-quenching activity. lqqE1 variant genes were generated by site-directed mutagenesis, where each residue was substituted by alanine (A). gfp, a negative control; momL, a positive control. F A LqqE1 structural modeling predicted by AlphaFold Protein Structure Database, and two key functions residue (E68, S126) shared in LqqE1 were shown in the predicted structure. In all panels, average data from three experiments was presented, ±SD. ***p < 0.0001, *p < 0.05, “ns” stands for not statistically significant, assessed by one-way ANOVA.