Figure 2.
Image Acquisition
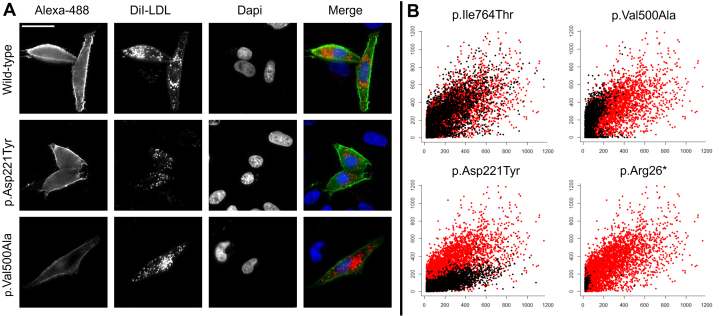
Cell-based functional profiling distinguishes functionally normal from functionally abnormal rare LDLR variants. (A) Images automatically acquired of Chinese hamster ovary (CHO)-ldlA7 cells transiently expressing wild-type LDLR, variant p.Asp221Tyr or variant p.Val500Ala. LDLR variants were characterized by monitoring cellular internalization of fluorescently labeled low-density lipoprotein (LDL) (DiI-LDL; red) and quantifying LDLR expression at cell surface (Alexa Fluor 488; green). Nuclei are labeled with 4′,6-diamidino-2-phenylindole (DAPI) (blue). All images were acquired on a fully automated widefield Olympus IX81 microscope, using an UPlanApo 20 × 0.7 NA objective and ScanR software (version 2.1.0.15, Olympus Biosciences). Bar = 25 μm. (B) Graphs depict relative signal intensities of total DiI-LDL in endocytic subcellular compartments (y-axis, in arbitrary units) plotted against LDLR expression (x-axis, in arbitrary units). Each individual scatter plot represents a LDLR variant (black dots) and the wild type (red dots), where each dot corresponds to a single cell. CHO-ldlA7 cells transiently expressing normal variant p.Ile764Thr (upper left), class II variant Val500Ala (upper right), class III variant p.Asp221Tyr (bottom left), and class I variant p.Arg26∗ (bottom right).