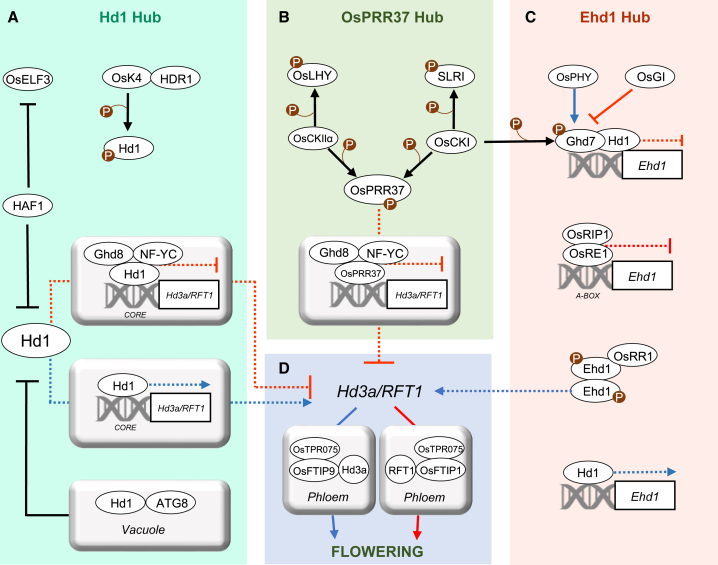
Figure 3.
Post-transcriptional levels of regulation in the flowering-time network.
We identified four hubs corresponding to Hd1, PRR37, Ehd1, and the florigens.
(A) Hd1 hub. Hd1 forms Hd1/NF-Y complexes that directly repress expression of florigens under LDs. Repression is released in SDs, and Hd1 becomes an activator. Hd1 stability depends on HAF1 and on components of the autophagy pathway, including ATG proteins, in the vacuole. Hd1 can be phosphorylated by OsK4, and this modification might impact Hd1 stability.
(B) OsPRR37 hub. OsPRR37 can replace Hd1 in an NF-Y complex and repress florigen expression under LDs. It can be phosphorylated by CKI and CKIIα. Phosphorylation might affect PRR37 stability or activity.
(C) Ehd1 hub. Ehd1 is repressed under LDs by the Ghd7/Hd1 and OsRE1/OsRIP1 complexes. Phosphorylation is essential for Ehd1 dimerization and activity. OsRR1 interacts with Ehd1 to form an inactive complex and inhibit its capacity to induce expression of the florigens. Phosphorylation of Ghd7 by CKI enhances its repressor activity.
(D) Florigen hub. Activity of the florigens depends on their transport in the phloem, which takes place by physical interaction with OsFTIP proteins and OsTPR075. Proteins are indicated by ovals and genes by rectangles. Names of DNA motifs bound by proteins or protein complexes are indicated below the double helix. Red and blue arrows indicate LD and SD regulation, respectively. Dashed arrows/flat-ended arrows indicate transcriptional activation/repression. Continuous arrows +P indicate phosphorylation. Continuous flat-ended arrows indicate protein degradation.