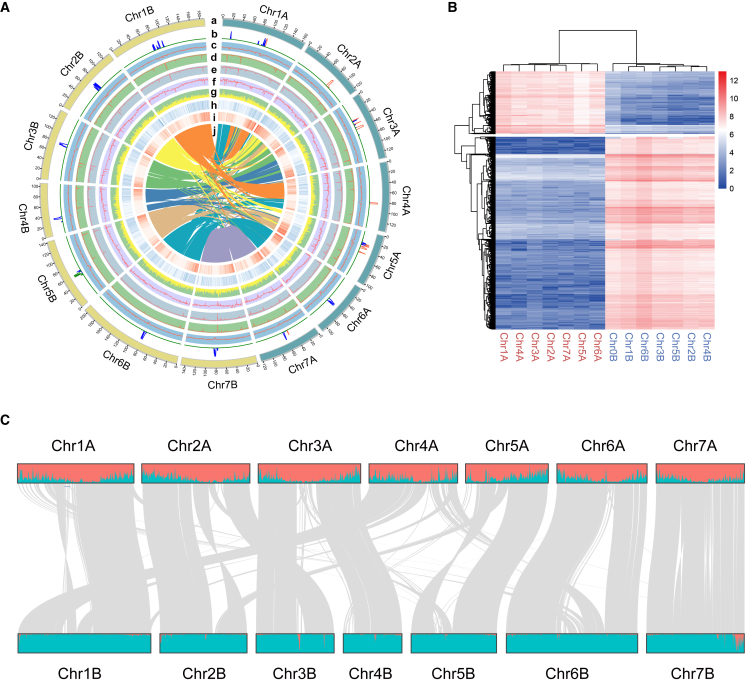
Figure 1.
Allotetraploid genome of JUJUNCAO (C. fungigraminus).
(A) Circos plot of the JUJUNCAO genome assembly. Quantitative tracks (b to j) are aggregated in 500-kbp bins, and independent y axis labels are as follows: (a) chromosomes; (b) centromeric repeat sequences; (c) GC content; (d) CG methylation; (e) CHG methylation; (f) CHH methylation; (g) LTRs; (H) SNPs; (h) gene expression; (i) gene density; (j) syntenic relationships.
(B) Clustering of counts of 13-mers that differentiate homeologous chromosomes enabled consistent partitioning of the genome into two subgenomes. Red and blue chromosome names correspond to the A and B subgenomes, respectively.
(C) Distribution of subgenome-specific 13-mer sequences (red for subgenome A and blue for subgenome B) for 14 chromosomes and synteny between the two subgenomes.