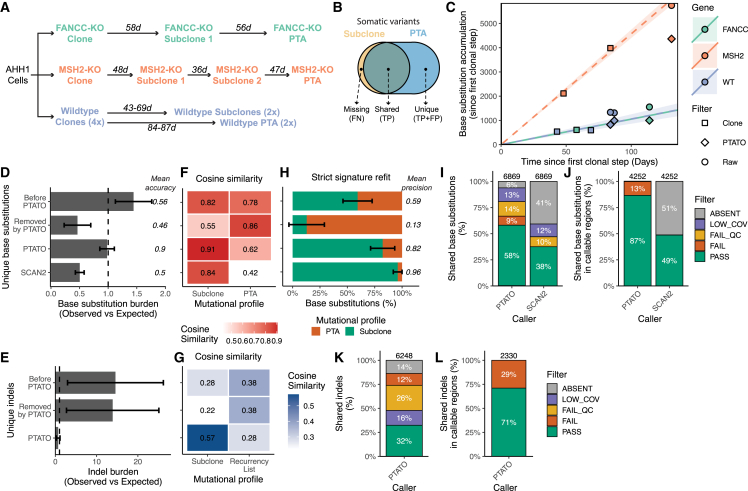
Figure 2.
Filtering by PTATO enables accurate analyses of somatic mutation patterns and burdens
(A) Schematic overview of the clonal steps performed for the three types of clonal cell lines generated in this study. Numbers indicate the days (d) in culture between the single-cell sorts, which are used to calculate mutation rates for each cell line.
(B) Venn diagram indicating which variants were used as false negatives (FN), true positives (TP), and false positives (FP).
(C) Accumulation of base substitutions per sample since the first clonal step. The circles and diamonds indicate the number of base substitutions detected in the PTA samples before and after PTATO filtering, respectively.
(D) Observed versus expected number of base substitutions in the PTA samples before PTATO filtering, removed by PTATO, after filtering by PTATO and after filtering by SCAN2. Data are represented as the mean (± SEM) in the four PTA samples.
(E) Observed versus expected (OE) number of indels in the PTA samples before or after filtering by PTATO and after filtering by SCAN2. Data are represented as the mean (± SEM) in the four PTA samples. Accuracy is determined as the mean absolute difference between the OE values and an OE value of 1.
(F) Heatmap showing the mean cosine similarities between the 96-trinucleotide profiles of the unique base substitutions before PTATO filtering, removed by PTATO, after PTATO filtering, or after SCAN2 calling and the profiles of the subclones analyzed by bulk WGS or the previously defined universal PTA artifact signature.18.
(G) Heatmap showing the mean cosine similarities between the profiles of the unique indels before PTATO filtering, removed by PTATO, or after PTATO filtering and the indel profiles of the subclones analyzed by bulk WGS or the list of recurrent indels used for filtering.
(H) Mean contributions (± SEM) of the universal PTA artifact signature and the mutational signatures of the subclones to the mutational profiles in the four PTA samples before PTATO filtering, removed by PTATO, after filtering by PTATO, or after filtering by SCAN2. Precision is determined as the mean contribution of the mutational signatures of the subclones to the mutational profiles of the PTA samples.
(I) Fractions of shared base substitutions present in the subclones that are also detected (PASS) in the PTA samples originating from these subclones by PTATO or SCAN2 (SCAN2 could not be used to study indels in these samples).
(J) Fractions of base substitutions after excluding the variants (in both the PTATO and SCAN2 call sets) with low coverage (LOW_COV), low genotype quality (LOW_QC), or undetected variants (ABSENT) as determined by PTATO. Few shared variants are (mis)classified as artifact (FAIL) in the PTA samples.
(K) Fractions of shared indels present in the subclones that are also detected (PASS) in the PTA samples originating from these subclones by PTATO or SCAN2 (SCAN2 could not be used to study indels in these samples).
(L) Fractions of indels after excluding the variants with low coverage (LOW_COV), low genotype quality (LOW_QC), or undetected variants (ABSENT) as determined by PTATO. Some indels are (mis)classified as artifact (FAIL) in the PTA samples (because they are present in the exclusion list or are insertions in long homopolymers).