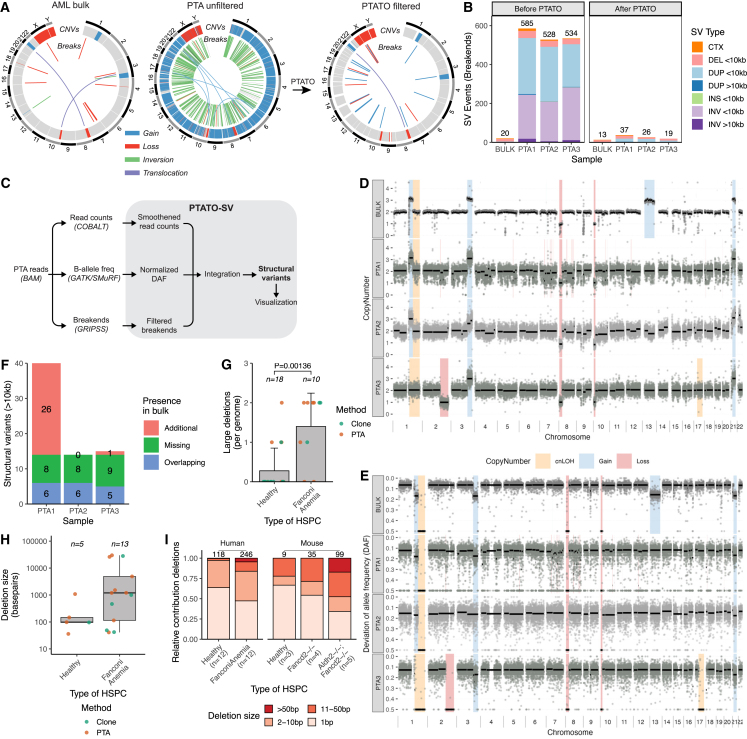
Figure 5.
SV filtering by PTATO reveals an increased deletion burden in HSPCs of patients with FA
(A) Circos plots showing copy number variants (CNVs) and balanced SVs in a PTA (left/center) and bulk WGS sample (right) of patient IBFM35. The standard SV calling pipeline for bulk WGS generates hundreds of false-positive calls in PTA samples (left), most of which are removed by PTATO filtering (center), leading to similar SV profiles as a sample sequenced by bulk WGS (right panel).
(B) Number of SV events detected by GRIDSS without filtering by PTATO (left) and the number of SVs remaining after filtering by PTATO (right) in bulk and PTA-based WGS samples of IBFM35.
(C) Schematic overview of the SV calling and filtering strategy tailored for PTA-based WGS data implemented in the PTATO pipeline.
(D) Copy number profiles (100-kb windows) of the AML bulk sample analyzed by the bulk WGS SV calling pipeline and three PTA samples analyzed by PTATO. Background shadings indicate the final copy number call made by PTATO (for PTA samples) or PURPLE (for the bulk WGS sample).
(E) Deviation of allele frequency (DAF) plots (100-kb windows) of the AML bulk sample and three PTA samples. The DAF depicts the absolute difference between 0.5 (perfect heterozygosity) and the actual allele frequency of a germline variant.
(F) Number of SVs (>10 kb in size) that are present in the HSPCs and present (“Overlapping”) or absent (“Additional”) in the AML bulk or present in the bulk but absent in the HSPCs (“Missing”).
(G) Number of deletions (>25 bp) detected by GRIDSS and PTATO in genomes of HSPCs of FA patients or healthy donors (including five cord blood samples sequenced after PTA). Numbers shown above the bars indicate the number of individuals per group. The p value was calculated by Wilcoxon Mann-Whitney test.
(H) Size (in bp) of each detected deletion in HSPCs of healthy donors and patients with FA (no significant difference Wilcoxon Mann-Whitney test). Numbers above the boxes indicate the total number of deletions per group.
(I) Distribution of the sizes of small (detected by GATK for the human samples) and large (detected by GRIDSS for the human samples) deletions in human and mice12 HSPCs with different genetic backgrounds. The numbers above the bars indicate the total number of deletions analyzed per group.