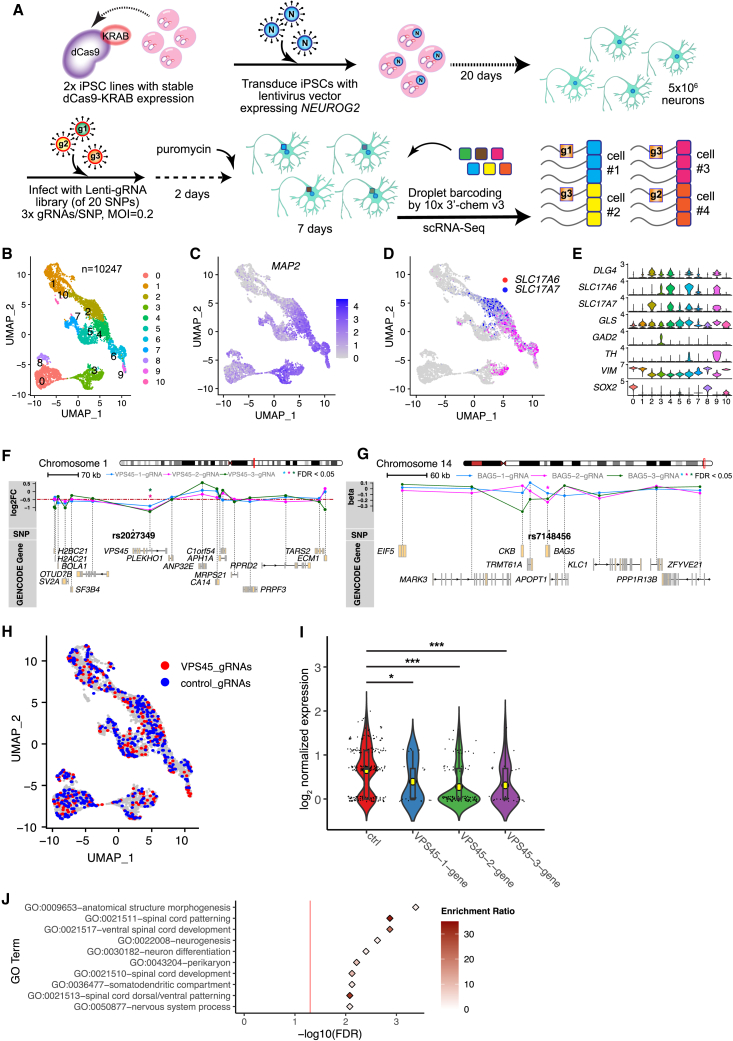
Figure 2.
Multiplex CRISPRi combined with scRNA-seq in NGN2-Glut identifies regulatory sequences flanking SZ-associated ASoC SNPs
(A) Modified CROP-seq approach to screen cis-targets of the 20 SNP sites in NGN2-Glut. Two iPSC lines (CD0000009, CD0000011) were used.
(B) Uniform Manifold Approximation and Projection (UMAP) plot showing the 11 clusters of the 10,247 cells used in the scRNA-seq analysis.
(C) UMAP plot showing the normalized expression of MAP2.
(D) UMAP plot showing the expression pattern of glutamatergic markers SLC17A7 (vGlut1) and SLC17A6 (vGlut2).
(E) Violin plots showing expression of neuron-specific markers.
(F) Gene track-expression plot showing the cis effects of CROP-seq gRNAs targeting rs2027349 (VPS45 site). ∗FDR <0.05.
(G) Same as (F), but for three CROP-seq gRNAs targeting rs7148456 (BAG5 site).
(H) UMAP plot showing cells assigned to one of the three VPS45 gRNAs (red) or control gRNAs (targeting GFP; blue).
(I) The effects of VPS45 gRNAs on the expression level of VPS45. Kruskal-Wallis test (non-parametric) was used (shown are Dunn’s multiple comparisons adjusted p; ∗∗∗p < 0.001).
(J) Top 10 enriched GO terms of genes up- or downregulated by ASoC-targeting gRNA-2 at the rs2027349 locus (VPS45). Red line, FDR 0.05.