Figure 6.
VPS45, AC244033.2 (lncRNA), and C1orf54 interactively altered neural phenotypes in NGN2-Glut at the rs2027349 locus
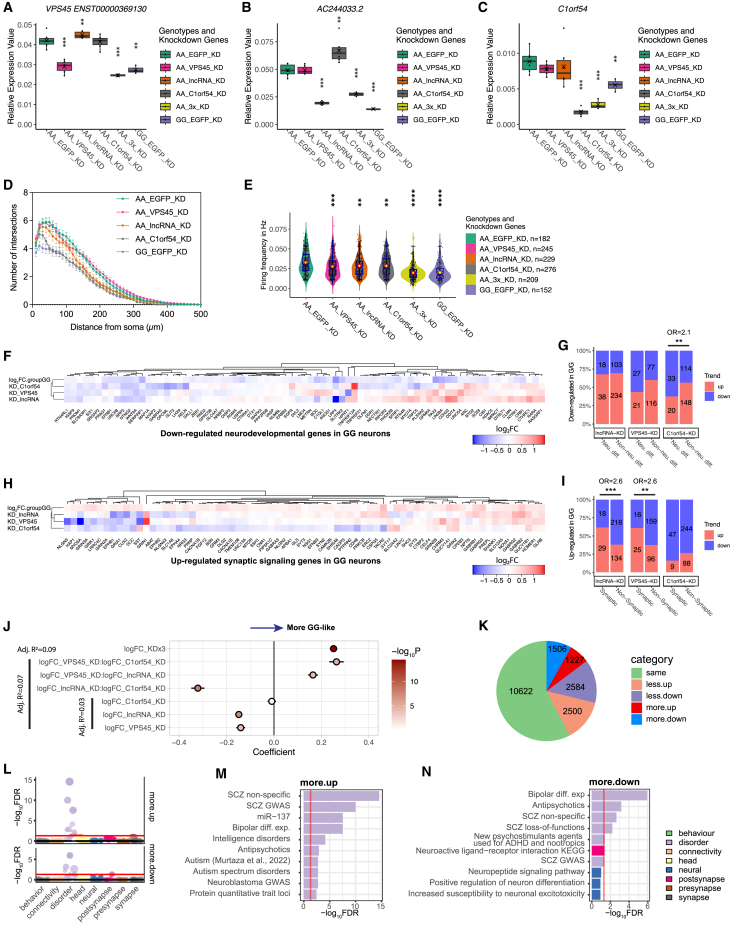
(A–C) Box-whisker plots showing that shRNA-mediated single and triple KD in AA neurons reversed the gene expression patterns to GG neurons. GAPDH was the endogenous control for normalization in qPCR assay. AA_EGFP_KD was used as the control for all. Saltire mark indicates the mean. Kruskal-Wallis test (non-parametric). For individual KD, two clones per donor line (CD0000011) and three biological replicates per clone were used. For triple KD, three biological replicates from one clone were used.
(D) Sholl analysis of individual KDs; refer to Figure S7B for statistics. Two clones per genotype from two independent experiments were used.
(E) Calcium imaging analysis of neuron firing frequency for gene KD. Kruskal-Wallis (non-parametric) test with Dunn’s multiple comparisons and adjusted p values. n = 181, 245, 229, 276, and 152 neurons. One or two clones per genotype from two independent experiments were used. ∗∗∗∗p < 0.0001.
(F) Hierarchical clustering of log2FC for neurodevelopmental genes downregulated in rs2027349-edited GG neurons and their expression changes in each shRNA KD in AA neurons.
(G) Enrichment of the up-/downregulated genes in each shRNA KD in AA neurons among the neurodevelopmental (i.e., neuron differentiation related) genes downregulated in rs2027349-edited GG neurons.
(H) Same as (F), using synaptic genes upregulated in GG neurons.
(I) Same as (G), examining the enrichment among synaptic genes upregulated in GG neurons. In (G) and (I), Fisher’s exact test (two-tailed) was used to estimate the enrichment; OR, odds ratio, ∗∗∗p < 0.001.
(J) Summary of the correlations from linear regression model fitting. Whiskers, ±95% CI; fill, −log10p.
(K) Pie chart showing the number and proportions of genes in each synergistic category in the combinatorial vs. the additive model.
(L) Competitive GSEA of DEGs in “more upregulated” (more.up) and “more downregulated” (more.down) categories based on 698 curated neural gene sets and stratified by eight neural categories.
(M and N) Bar plots of overrepresentation analysis in (L) using a hypergeometric test of the gene sets at FDR <0.05 and ranked by FDR value. Red lines, FDR 0.05.