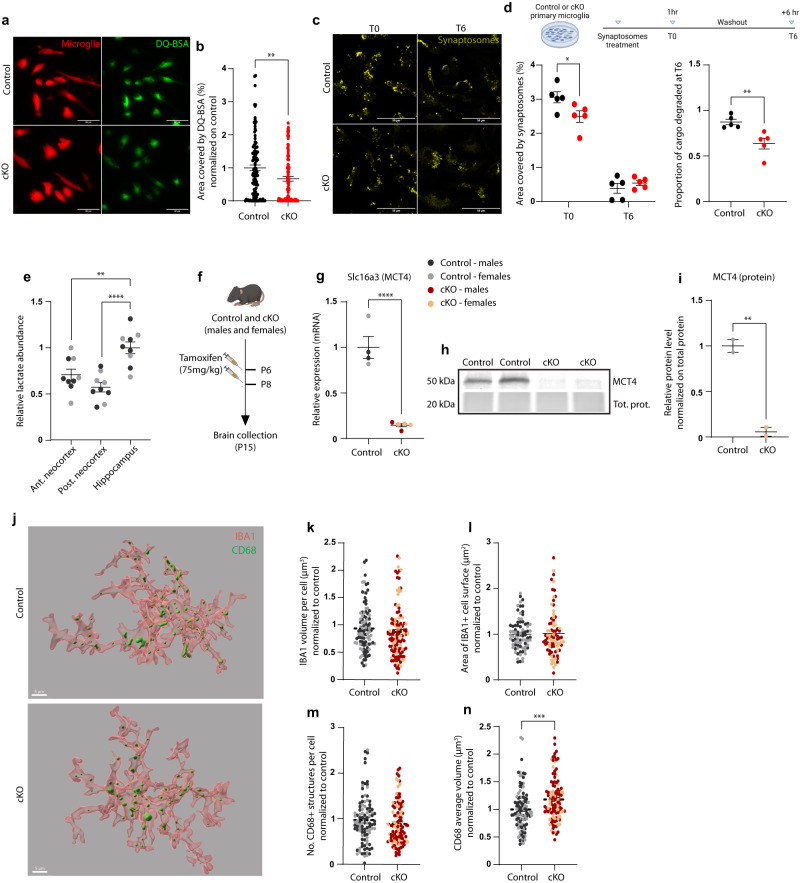
Fig. 3. MCT4 cKO microglia display reduced phagocytosis and lysosomal dysfunction in vitro and alteration of phagolysosomal structures in vivo.
a Representative z-stack projections of microglia (TdTomato+) treated with DQ-BSA, and b relative quantification of area covered by cleaved DQ-BSA in control and cKO cells. Data point represent individual cells, control: n = 111 cells; cKO: n = 103 cells from N = 2 independent experiments. Two-tailed unpaired t test, **p = 0.0036; Scale bar: 50 µm. c Representative confocal z-stack acquisitions of primary microglia exposed to fluorescently labeled synaptosomes (scale bar: 50 µm), and d relative quantification. Experiment timeline created with BioRender.com. Left: Microglial area covered by fluorescent signal per cell after 1 h exposure to synaptosomes (T0) and 6 h after medium washout (T6). Two-way ANOVA followed by Sidak’s post hoc multiple comparison test, *p = 0.0248. Right: proportion of cargo degraded in 6 h in control and cKO cells. Two-tailed unpaired t test, **p = 0.0074. Data points represent the average of at least 30 cells per condition, from N = 5 independent experiments. e Relative lactate abundance measured by LC-MS in different brain areas of P15 control mice. N = 5 males (black dots) and N = 4 females (gray dots). One-way ANOVA followed by Dunnett’s post hoc multiple comparison test; hippocampus vs. anterior neocortex: **p = 0.0033; hippocampus vs. posterior neocortex: ****p < 0.0001. f Schematic of experimental design for tamoxifen treatment and brain collection at P15. Created with BioRender.com. g Expression of Slc16a3 mRNA and h, i MCT4 protein in microglia acutely isolated from control and cKO mice at P15; for mRNA: Control: N = 4 (N = 2 males and N = 2 females) vs. cKO: N = 5 (N = 2 males and N = 3 females). For protein: N = 2 control females and N = 2 cKO females). Two-tailed unpaired t test, ****p < 0.0001, **p = 0.0082. j Representative 3D reconstruction of microglial cells from the CA1 stratum radiatum of control and cKO mice at P15, scale bar: 5 µm. Quantification of IBA1+ k cell volume, l surface area, m number of CD68+ structures within each microglia cell, and n average volume of CD68+ structures per cell, normalized to controls. k–n Data points represent single cells from N = 4 males and N = 4 females per genotype (P15); control: n = 79–103 cells; cKO: n = 83–113 cells. Two-tailed unpaired t test, ***p = 0.0007. b, d, e, g, i, k–n Data are represented as mean ± SEM. Source data are provided as a Source Data file.