Abstract
X-ray crystallography and X-ray spectroscopy using X-ray free electron lasers plays an important role in understanding the interplay of structural changes in the protein and the chemical changes at the metal active site of metalloenzymes through their catalytic cycles. As a part of such an effort, we report here our recent development of methods for X-ray absorption spectroscopy (XAS) at XFELs to study dilute biological samples, available in limited volumes. Our prime target is Photosystem II (PS II), a multi subunit membrane protein complex, that catalyzes the light-driven water oxidation reaction at the Mn4CaO5 cluster. This is an ideal system to investigate how to control multi-electron/proton chemistry, using the flexibility of metal redox states, in coordination with the protein and the water network. We describe the method that we have developed to collect XAS data using PS II samples with a Mn concentration of <1 mM, using a drop-on-demand sample delivery method.
Keywords: Manganese, photosystem II, PhotoIUPAC 2022, water oxidation, X-ray absorption spectroscopy, X-ray free electron lasers
Introduction
Nature uses remarkably varied systems and mechanisms to perform complex chemical transformations with efficiency, speed, and specificity. At the active site of many enzymes are metal centers, responsible for the rearrangement of electrons, protons, and atoms to carry out electron transfer and catalytic reactions [1]. X-ray crystallography and spectroscopy at synchrotrons has been intensively used to understand the function of metalloenzymes, and it has produced a wealth of valuable information from many different systems. These experiments are generally carried out in synchrotron facilities at cryogenic temperature to minimize radiation-induced changes by reducing the diffusion rate of solvated electrons and hydroxyl radicals. The introduction of X-ray free electron lasers (XFELs) over the last decade has opened up the possibility to apply X-ray techniques at room temperature through the ability to collect data before the onset of radiation-induced sample damage [2]. Such ‘operando’ characterization of biological samples has been demonstrated to be a powerful approach to capture catalytic intermediates and chemical and structural sequences of reactions. In addition, shot-by-shot data collection with very intense and ultrashort (10–40 fs) X-ray pulses at XFELs enabled the simultaneous collection of X-ray crystallography and X-ray emission spectroscopy (XES) data using micron-sized single crystal suspensions [3]. This methodology has provided information about the interplay between the metal catalytic centers and their environment from the same sample, and many successful applications are reported in the recent literature [3], [4], [5], [6], [7].
Another important technique that has been widely used in the field of metalloenzymes is X-ray Absorption Spectroscopy (XAS) [8]. Transition metal K-edge XAS has been used primarily in two ways: for studying (1) the oxidation states and electronic structure of metal and ligand atoms using X-ray absorption near edge spectroscopy (XANES), and (2) the local coordination environment of metals and atomic distances using extended X-ray absorption fine structure (EXAFS). For both crystalline and non-crystalline samples, EXAFS has been a tool of choice to determine the metal-metal distances in multi-metallic centers and metal-ligand bond distances with a sensitivity (<0.1 Å) surpassing what is generally possible from diffraction methods. X-ray absorption spectroscopy is therefore highly beneficial for the study of metalloenzymes using time-resolved studies of the metal catalytic centers to capture electronic and structural changes.
Unlike XES (which has no specific requirements on the excitation energy except that it needs to be above the absorption edge of the metal studied), XAS is less widely used at XFELs, as it involves scanning the incident X-ray energy over a wide range (<50 eV for XANES and >50 eV for EXAFS), while accurately normalizing signals from one pulse to another and at different incident energies. Intrinsically, XFEL pulses are stochastic with complex spectral and temporal pulse profiles and intensity fluctuations. There are additional fluctuations of the probed sample volume as it is replaced for every pulse, requiring new methods for normalizing the data as compared to those used at synchrotron radiation (SR) sources. Furthermore, the incident flux is reduced by a factor of about 100 when monochromatizing the self-amplified spontaneous emission (SASE) X-ray pulses, which have a relative bandwidth (ΔE) of ΔE/E 0 ≈ 0.5 % (E 0: incident X-ray energy, eV). Despite these difficulties, there have been successful XAS studies at XFELs [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]. Our previous report demonstrated that it is feasible to collect XAS data from ∼10 mM metal aqueous solution samples when normalizing through the use of the scattering signal from water collected on a downstream detector [9]. This method showed a linear relationship of the signal intensity with the probed sample volume, which can vary shot by shot, and the intensity of the X-ray pulses per shot.
In order to realistically collect XAS data from biological samples, one must overcome another major barrier, that is to decrease sample consumption. Biological samples are often hard to purify and produce in large quantities, which limits the amount of sample available for data collection. Additionally, one cannot often achieve a high concentration of proteins; for example, a maximum concentration of Mn in a typical solution Photosystem II (PS II) sample, which we describe in the following section, is ∼0.5 mM. We have developed an efficient drop-on-demand sample delivery system which requires minimum volume of samples. Strides have also been made utilizing microdrops and electrospinning jets, but these will not be discussed here [12, 22, 23]. Below we illustrate the use of the drop-on-demand method for XAS data collection using PS II samples.
Manganese cluster in PS II
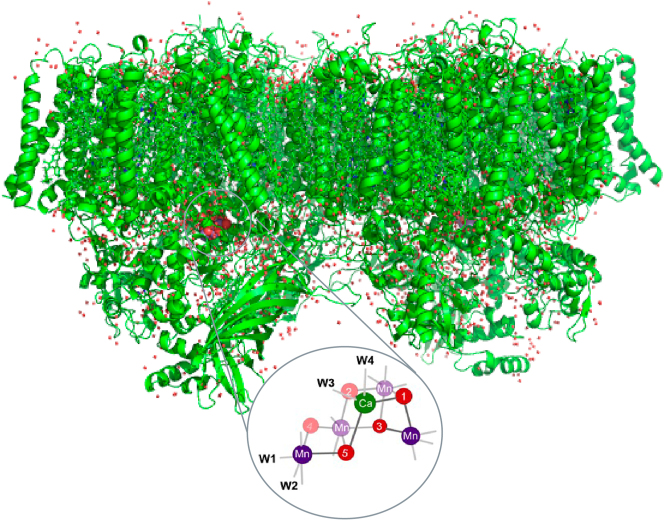
In PS II, which is shown in Fig. 1, four photons are used for charge separation at the reaction center and their photochemical energy is used for splitting two water molecules into molecular oxygen, four protons, and four electrons. During the reaction, four Mn atoms in the Mn4CaO5 cluster advance through redox changes as they cycle through the four intermediate states, S1, S2, S3 and S0. The S1 state is the dark resting state which is formally Mn(III)2Mn(IV)2. During the S1 to S2 and the S2 to S3 transitions, Mn goes from Mn(III) to Mn(IV) in each step, and the accumulated oxidizing power is released during the S3 to S0 transition to form an O–O bond and the release of O2. Over the last 10 years, serial crystallography using XFELs has provided insight into the structures of these intermediates states [24], [25], [26] as well as some of the chemical processes occurring, including the oxidation kinetics of Mn using Mn Kß XES [4]. One of the fundamental questions, however, is the redox process during the S3 to S0 transition, where another oxidation of Mn (+IV to +V) or ligand (O·) occurs, followed by a four-electron reduction. A time-resolved XAS study at XFELs is expected to provide critical information on the mechanism of this step. In the following section, we used a drop-on-tape (DOT) sample delivery method [27] to achieve stable operation, and demonstrated the feasibility of the XFEL-based XAS method on dilute PS II samples at a Mn concentration of ∼0.5 mM.
Fig. 1:
PS II room temperature crystallographic structure in the dark stable S1 state [4], with the Mn cluster of the oxygen evolving complex (OEC) in the inset. The Mn cluster shows manganese (purple), bridging oxygens (red), calcium (green) and ligated waters (W1–W4). The formal oxidation state of the Mn atoms is Mn(III)2Mn(IV)2.
Experimental setup of the drop-on-tape XAS at XFELs
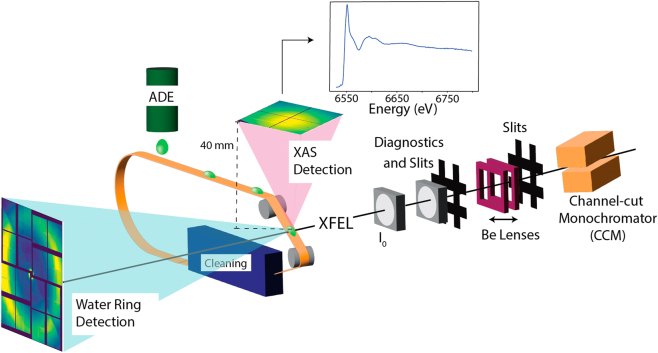
Figure 2 shows the DOT setup that is used for the XAS data collection. The DOT uses an acoustic droplet ejector (ADE) to produce 0.8–6 nL droplets, resulting in a solvent pathlength of 200–300 µm [27] at the X-ray interaction point. The droplets are formed from a reservoir that has a volume of 10 µL and is continuously refilled with sample via a syringe pump. These droplets are deposited on a Kapton tape that carries them into the path of the XFEL. The sample can be either solution or a crystal suspension. The total fluorescence signal is collected on an ePix-100 detector [28] 40 mm from the X-ray interaction point at an angle of 90° from the beam, to minimize scattering. With an active detector surface size of 39 mm × 40 mm, it covers a solid angle of 0.64 sr. In the forward direction, the water scattering signal (“water ring”) from the droplet is collected on an ePix-10k2m detector at a distance of 80 mm. This scattering signal is used for normalization of the total fluorescence XAS signal.
Fig. 2:
Schematic of the XAS setup using the DOT (Drop-on-Tape) method. Use of ADE (acoustic droplet ejector) to deposit samples in ∼250 μm droplets reduces sample consumption which facilitates the collection of data for <1 mM metal concentration in biological samples. Total fluorescence from the sample is collected with an ePix detector directly above the droplet from the droplet-XFEL interaction point. A forward scattering detector collects scattering from the solvent that is used for normalization. Figure expanded from original publication in Chatterjee et al. 2019 [9].
Unlike a liquid jet, DOT can deliver samples matched with the rate of the XFEL pulse frequency (120 Hz for LCLS) or lower frequency. This drastically reduces the amount of sample required for the experiment. For example, in Chatterjee et al. in order to obtain a spectrum from a sample of 10 mM Mn concentration with an XAS edge jump of 1,000,000 counts, the Rayleigh jet (500 μm diameter) consumed 525 mL (35 min at 120 Hz with 100 % hit rate), while the DOT (200 μm probed diameter of the droplets) consumed 5 mL (120 min at 120 Hz with 80 % hit rate) [9].
XAS data of PS II
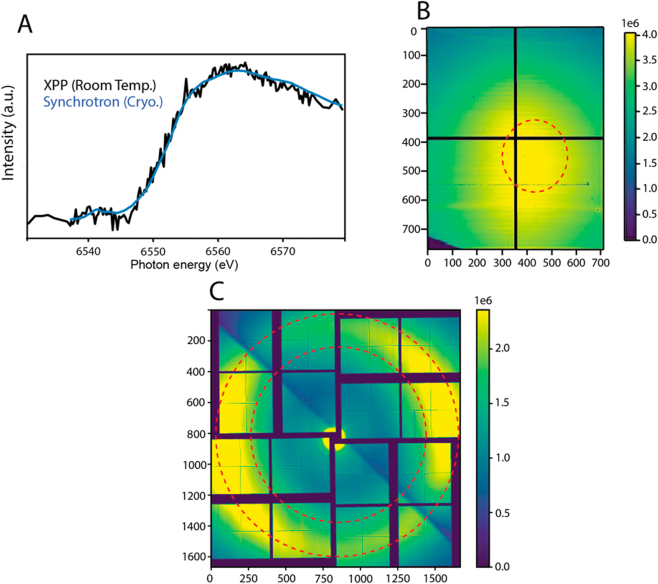
Room temperature XAS data of the PS II dark S1 state is shown in Fig. 3A. The data was collected at the Linac Coherent Light Source (LCLS) using the X-ray pump and probe (XPP) beamline [29]. The SASE beam had a pulse length of ∼30 fs, bandwidth of 20 eV, and a pulse energy of 1.8 mJ before the monochromator corresponding to 1.7 × 1012 photons per pulse, at a repetition rate of 120 Hz. The beam focus was 3 μm full width half maximum (FWHM). SASE beam was used over seeded beam (typically 0.4–1 eV), because the seeded beam only offers a minor increase in photons and requires significant machine tuning. A more complete discussion is found in Chatterjee et al. [9].
Fig. 3:
(A) Room temperature Mn K edge XAS spectrum from dark stable S1 state PS II (Black) collected at XPP (LCLS), in comparison to a corresponding synchrotron spectrum (blue) reported previously [30]. LCLS XAS data was collected from PS II solution sample (0.5 mM Mn) in 35 min with 120 Hz at XPP. Sample flow rate into the reservoir from which droplets are produced was 48 μL/min, and the total consumed sample volume was 1.6 mL. (B) Cumulative total fluorescence ePix-100 detector signal for XAS. A red dotted circle shows the region of interest (ROI). (C) Water scattering on the ePix-10k2m detector; the region used for normalization is located between the two red circles.
The XPP Si(111) channel-cut monochromator (CCM), with a resolution of 1.4 × 10−4 ΔE/E, was used to monochromatize the incoming X-ray pulses, yielding <1 % of the SASE flux (∼1 × 1010 photons per pulse) in the beam after the monochromator. By changing the CCM angle, the X-ray energy was scanned, ensuring roughly equal photon numbers at each energy point over the chosen scan range. Three beryllium compound refractive lenses (thicknesses of 100, 200, and 500 μm) with a focal spot of 4.0 m were used to maintain focus while scanning, and the center of the SASE bandwidth followed the monochromator scanning energy by adjusting the electron beam energy. Post monochromator beam intensity was recorded for each shot using one of the XPP I 0 intensity position monitors that measure the intensity of the X-rays scattered from a thin foil inserted into the beam. The scan protocol was 6530–6535 eV (1 eV intervals), 6535–6575 eV (0.25 eV intervals), and 6575–6580 eV (1 eV intervals) with an exposure time of 1 s per energy. The monochromator was scanned step-by-step as defined by the scan protocol. However, the data collection was continuous, and therefore an occasional shot occurs during the monochromator movement. One scan, consisting of both the forward scan and the backward scan through the chosen energy range took 7 min. The monochromator was calibrated using a Mn foil edge energy of 6555.8 eV. Sample flow rate into the droplet dispenser reservoir was 48 μL/min and droplets were generated at a frequency of 120 Hz. The sample was an aqueous solution of PS II isolated from the thermophilic cyanobacterium Thermosynechococus vestitus at a chlorophyll concentration of 4.2 mM, corresponding to ∼0.5 mM Mn.
An ePix-100 detector (50 µm pixel, 768 × 704 pixels) was used to collect the XAS spectra in total fluorescence mode, and an ePix-10k2m detector (1662 × 1663 pixels) was used to collect the solvent scattering signal (“water ring”) for normalization. The selected region of interest (ROI) for both detectors is shown in Fig. 3B,C. The ROI for total fluorescence was the region of highest fluorescence photon yield with minimal scatter (Fig. 3B). The solvent scatter was selected through an azimuthal average which integrated radially between two defined radii from the center of the detector. These radii corresponded to the inner and outer edge of the solvent scattering ring (shown between the red lines, Fig. 3C). Shots with a water ring integration value (WIV) of less than 500 a.d.u. were considered misses and removed from the data set. The WIV was used to normalize fluorescence data; accounting for variation of droplet size, hit rate, and XFEL pulse intensity on a shot-by-shot basis. Previous studies have proven this method to be superior to normalization done by XPP I 0 intensity position monitors [9].
In Fig. 3A, the XANES spectrum of the PS II dark stable S1 state is compared with the synchrotron data collected at cryogenic temperature (∼8 K) [30]. The XFEL data show similar edge locations and shapes as the synchrotron data. The spectrum was collected in 35 min with a sample consumption of 1.6 mL.
Outlook
The current study demonstrated the feasibility of XAS data collection at an XFEL using a limited volume of biological samples at sub mM concentrations. We show that a room temperature XANES spectrum of a sample with ∼0.5 mM metal concentration can be collected from ∼1.6 mL volume in 35 min using the current setup. Further developments are on the way to reduce the background signal that goes into the XAS detector, which will further reduce the sample volume and time required for XAS data collection. Furthermore, this will also help enable EXAFS data collection of biological samples at XFELs. These advances will bring XFELs into the mainstream of XANES and EXAFS studies of metalloenzymes which are now primarily conducted at synchrotron radiation sources.
Acknowledgement
This work was supported by the Director, Office of Science, Office of Basic Energy Sciences (OBES), Division of Chemical Sciences, Geosciences, and Biosciences (CSGB) of the Department of Energy (DOE) (J.Y., V.K.Y., J.K.) for X-ray spectroscopy data collection and analysis and methods development for photosynthetic systems, by the National Institutes of Health (NIH) Grants GM055302 (V.K.Y.) for PS II biochemistry, GM110501 (J.Y.) and GM126289 (J.K.) for instrumentation development for XFEL experiments. Use of the LCLS and SSRL, SLAC National Accelerator Laboratory, is supported by the U.S. DOE, Office of Science, OBES under Contract No. DE-AC02-76SF00515, and X-ray spectroscopy work at the LCLS is supported by NIH grant P41GM139687. We thank the support staff at LCLS/SLAC and SSRL.
References
- [1].Holm R. H., Solomon E. I. Chem. Rev. . 2014;114:3367. doi: 10.1021/cr500118g. [DOI] [PubMed] [Google Scholar]
- [2].Bergmann U., Kern J., Schoenlein R. W., Wernet P., Yachandra V. K., Yano J. Nat. Rev. Phys. . 2021;3:264. doi: 10.1038/s42254-021-00289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kern J., Alonso-Mori R., Tran R., Hattne J., Gildea R. J., Echols N., Glöckner C., Hellmich J., Laksmono H., Sierra R. G., Lassalle-Kaiser B., Koroidov S., Lampe A., Han G., Gul S., DiFiore D., Milathianaki D., Fry A. R., Miahnahri A., Schafer D. W., Messerschmidt M., Seibert M. M., Koglin J. E., Sokaras D., Weng T.-C., Sellberg J., Latimer M. J., Grosse-Kunstleve R. W., Zwart P. H., White W. E., Glatzel P., Adams P. D., Bogan M. J., Williams G. J., Boutet S., Messinger J., Zouni A., Sauter N. K., Yachandra V. K., Bergmann U., Yano J. Science . 2013;340:491. doi: 10.1126/science.1234273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ibrahim M., Fransson T., Chatterjee R., Cheah M. H., Hussein R., Lassalle L., Sutherlin K. D., Young I. D., Fuller F. D., Gul S., Kim I.-S., Simon P. S., de Lichtenberg C., Chernev P., Bogacz I., Pham C. C., Orville A. M., Saichek N., Northen T., Batyuk A., Carbajo S., Alonso-Mori R., Tono K., Owada S., Bhowmick A., Bolotovsky R., Mendez D., Moriarty N. W., Holton J. M., Dobbek H., Brewster A. S., Adams P. D., Sauter N. K., Bergmann U., Zouni A., Messinger J., Kern J., Yachandra V. K., Yano J. Proc. Natl. Acad. Sci. USA . 2020;117:12624. doi: 10.1073/pnas.2000529117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rabe P., Kamps J. J. A. G., Sutherlin K. D., Linyard J. D. S., Aller P., Pham C. C., Makita H., Clifton I., McDonough M. A., Leissing T. M., Shutin D., Lang P. A., Butryn A., Brem J., Gul S., Fuller F. D., Kim I.-S., Cheah M. H., Fransson T., Bhowmick A., Young I. D., O’Riordan L., Brewster A. S., Pettinati I., Doyle M., Joti Y., Owada S., Tono K., Batyuk A., Hunter M. S., Alonso-Mori R., Bergmann U., Owen R. L., Sauter N. K., Claridge T. D. W., Robinson C. V., Yachandra V. K., Yano J., Kern J. F., Orville A. M., Schofield C. J. Sci. Adv. . 2021;7:eabh0250. doi: 10.1126/sciadv.abh0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Srinivas V., Banerjee R., Lebrette H., Jones J. C., Aurelius O., Kim I.-S., Pham C. C., Gul S., Sutherlin K. D., Bhowmick A., John J., Bozkurt E., Fransson T., Aller P., Butryn A., Bogacz I., Simon P., Keable S., Britz A., Tono K., Kim K. S., Park S.-Y., Lee S. J., Park J., Alonso-Mori R., Fuller F. D., Batyuk A., Brewster A. S., Bergmann U., Sauter N. K., Orville A. M., Yachandra V. K., Yano J., Lipscomb J. D., Kern J., Högbom M. J. Am. Chem. Soc. . 2020;142:14249. doi: 10.1021/jacs.0c05613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ohmer C. J., Dasgupta M., Patwardhan A., Bogacz I., Kaminsky C., Doyle M. D., Chen P. Y.-T., Keable S. M., Makita H., Simon P. S., Massad R., Fransson T., Chatterjee R., Bhowmick A., Paley D. W., Moriarty N. W., Brewster A. S., Gee L. B., Alonso-Mori R., Moss F., Fuller F. D., Batyuk A., Sauter N. K., Bergmann U., Drennan C. L., Yachandra V. K., Yano J., Kern J. F., Ragsdale S. W. J. Inorg. Biochem. . 2022;230:111768. doi: 10.1016/j.jinorgbio.2022.111768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ward J., Ollmann E., Maxey E., Finney L. A. Methods Mol. Biol. Clifton NJ . 2014;1122:171. doi: 10.1007/978-1-62703-794-5_12. [DOI] [PubMed] [Google Scholar]
- [9].Chatterjee R., Weninger C., Loukianov A., Gul S., Fuller F. D., Cheah M. H., Fransson T., Pham C. C., Nelson S., Song S., Britz A., Messinger J., Bergmann U., Alonso-Mori R., Yachandra V. K., Kern J., Yano J. J. Synchrotron Radiat. . 2019;26:1716. doi: 10.1107/s1600577519007550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Miller N. A., Deb A., Alonso-Mori R., Garabato B. D., Glownia J. M., Kiefer L. M., Koralek J., Sikorski M., Spears K. G., Wiley T. E., Zhu D., Kozlowski P. M., Kubarych K. J., Penner-Hahn J. E., Sension R. J. J. Am. Chem. Soc. . 2017;139:1894. doi: 10.1021/jacs.6b11295. [DOI] [PubMed] [Google Scholar]
- [11].Miller N. A., Deb A., Alonso-Mori R., Glownia J. M., Kiefer L. M., Konar A., Michocki L. B., Sikorski M., Sofferman D. L., Song S., Toda M. J., Wiley T. E., Zhu D., Kozlowski P. M., Kubarych K. J., Penner-Hahn J. E., Sension R. J. J. Phys. Chem. A . 2018;122:4963. doi: 10.1021/acs.jpca.8b04223. [DOI] [PubMed] [Google Scholar]
- [12].Miller N. A., Michocki L. B., Alonso-Mori R., Britz A., Deb A., DePonte D. P., Glownia J. M., Kaneshiro A. K., Kieninger C., Koralek J., Meadows J. H., van Driel T. B., Kräutler B., Kubarych K. J., Penner-Hahn J. E., Sension R. J. J. Phys. Chem. Lett. . 2019;10:5484. doi: 10.1021/acs.jpclett.9b02202. [DOI] [PubMed] [Google Scholar]
- [13].Michocki L. B., Miller N. A., Alonso-Mori R., Britz A., Deb A., Glownia J. M., Kaneshiro A. K., Konar A., Koralek J., Meadows J. H., Sofferman D. L., Song S., Toda M. J., van Driel T. B., Kozlowski P. M., Kubarych K. J., Penner-Hahn J. E., Sension R. J. J. Phys. Chem. B . 2019;123:6042. doi: 10.1021/acs.jpcb.9b05854. [DOI] [PubMed] [Google Scholar]
- [14].Miller N. A., Michocki L. B., Konar A., Alonso-Mori R., Deb A., Glownia J. M., Sofferman D. L., Song S., Kozlowski P. M., Kubarych K. J., Penner-Hahn J. E., Sension R. J. J. Phys. Chem. B . 2020;124:199. doi: 10.1021/acs.jpcb.9b09286. [DOI] [PubMed] [Google Scholar]
- [15].Bacellar C., Kinschel D., Mancini G. F., Ingle R. A., Rouxel J., Cannelli O., Cirelli C., Knopp G., Szlachetko J., Lima F. A., Menzi S., Pamfilidis G., Kubicek K., Khakhulin D., Gawelda W., Rodriguez-Fernandez A., Biednov M., Bressler C., Arrell C. A., Johnson P. J. M., Milne C. J., Chergui M. Proc. Natl. Acad. Sci. USA . 2020;117:21914. doi: 10.1073/pnas.2009490117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shelby M. L., Lestrange P. J., Jackson N. E., Haldrup K., Mara M. W., Stickrath A. B., Zhu D., Lemke H. T., Chollet M., Hoffman B. M., Li X., Chen L. X. J. Am. Chem. Soc. . 2016;138:8752. doi: 10.1021/jacs.6b02176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Levantino M., Lemke H. T., Schirò G., Glownia M., Cupane A., Cammarata M. Struct. Dyn. . 2015;2:041713. doi: 10.1063/1.4921907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Katayama T., Inubushi Y., Obara Y., Sato T., Togashi T., Tono K., Hatsui T., Kameshima T., Bhattacharya A., Ogi Y., Kurahashi N., Misawa K., Suzuki T., Yabashi M. Appl. Phys. Lett. . 2013;103:131105. doi: 10.1063/1.4821108. [DOI] [Google Scholar]
- [19].Obara Y., Ito H., Ito T., Kurahashi N., Thürmer S., Tanaka H., Katayama T., Togashi T., Owada S., Yamamoto Y., Karashima S., Nishitani J., Yabashi M., Suzuki T., Misawa K. Struct. Dyn. . 2017;4:044033. doi: 10.1063/1.4989862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mara M. W., Hadt R. G., Reinhard M. E., Kroll T., Lim H., Hartsock R. W., Alonso-Mori R., Chollet M., Glownia J. M., Nelson S., Sokaras D., Kunnus K., Hodgson K. O., Hedman B., Bergmann U., Gaffney K. J., Solomon E. I. Science . 2017;356:1276. doi: 10.1126/science.aam6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Britz A., Abraham B., Biasin E., van Driel T. B., Gallo A., Garcia-Esparza A. T., Glownia J., Loukianov A., Nelson S., Reinhard M., Sokaras D., Alonso-Mori R. Phys. Chem. Chem. Phys. . 2020;22:2660. doi: 10.1039/c9cp03483h. [DOI] [PubMed] [Google Scholar]
- [22].Sierra R. G., Laksmono H., Kern J., Tran R., Hattne J., Alonso-Mori R., Lassalle-Kaiser B., Glöckner C., Hellmich J., Schafer D. W., Echols N., Gildea R. J., Grosse-Kunstleve R. W., Sellberg J., McQueen T. A., Fry A. R., Messerschmidt M. M., Miahnahri A., Seibert M. M., Hampton C. Y., Starodub D., Loh N. D., Sokaras D., Weng T.-C., Zwart P. H., Glatzel P., Milathianaki D., White W. E., Adams P. D., Williams G. J., Boutet S., Zouni A., Messinger J., Sauter N. K., Bergmann U., Yano J., Yachandra V. K., Bogan M. J. Acta Crystallogr. D Biol. Crystallogr. . 2012;68:1584. doi: 10.1107/s0907444912038152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sierra R. G., Gati C., Laksmono H., Dao E. H., Gul S., Fuller F., Kern J., Chatterjee R., Ibrahim M., Brewster A. S., Young I. D., Michels-Clark T., Aquila A., Liang M., Hunter M. S., Koglin J. E., Boutet S., Junco E. A., Hayes B., Bogan M. J., Hampton C. Y., Puglisi E. V., Sauter N. K., Stan C. A., Zouni A., Yano J., Yachandra V. K., Soltis S. M., Puglisi J. D., DeMirci H. Nat. Methods . 2016;13:59. doi: 10.1038/nmeth.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kern J., Chatterjee R., Young I. D., Fuller F. D., Lassalle L., Ibrahim M., Gul S., Fransson T., Brewster A. S., Alonso-Mori R., Hussein R., Zhang M., Douthit L., de Lichtenberg C., Cheah M. H., Shevela D., Wersig J., Seuffert I., Sokaras D., Pastor E., Weninger C., Kroll T., Sierra R. G., Aller P., Butryn A., Orville A. M., Liang M., Batyuk A., Koglin J. E., Carbajo S., Boutet S., Moriarty N. W., Holton J. M., Dobbek H., Adams P. D., Bergmann U., Sauter N. K., Zouni A., Messinger J., Yano J., Yachandra V. K. Nature . 2018;563:421. doi: 10.1038/s41586-018-0681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hussein R., Ibrahim M., Bhowmick A., Simon P. S., Chatterjee R., Lassalle L., Doyle M., Bogacz I., Kim I.-S., Cheah M. H., Gul S., de Lichtenberg C., Chernev P., Pham C. C., Young I. D., Carbajo S., Fuller F. D., Alonso-Mori R., Batyuk A., Sutherlin K. D., Brewster A. S., Bolotovsky R., Mendez D., Holton J. M., Moriarty N. W., Adams P. D., Bergmann U., Sauter N. K., Dobbek H., Messinger J., Zouni A., Kern J., Yachandra V. K., Yano J. Nat. Commun. . 2021;12:6531. doi: 10.1038/s41467-021-26781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Suga M., Akita F., Yamashita K., Nakajima Y., Ueno G., Li H., Yamane T., Hirata K., Umena Y., Yonekura S., Yu L.-J., Murakami H., Nomura T., Kimura T., Kubo M., Baba S., Kumasaka T., Tono K., Yabashi M., Isobe H., Yamaguchi K., Yamamoto M., Ago H., Shen J.-R. Science . 2019;366:334. doi: 10.1126/science.aax6998. [DOI] [PubMed] [Google Scholar]
- [27].Fuller F. D., Gul S., Chatterjee R., Burgie E. S., Young I. D., Lebrette H., Srinivas V., Brewster A. S., Michels-Clark T., Clinger J. A., Andi B., Ibrahim M., Pastor E., de Lichtenberg C., Hussein R., Pollock C. J., Zhang M., Stan C. A., Kroll T., Fransson T., Weninger C., Kubin M., Aller P., Lassalle L., Bräuer P., Miller M. D., Amin M., Koroidov S., Roessler C. G., Allaire M., Sierra R. G., Docker P. T., Glownia J. M., Nelson S., Koglin J. E., Zhu D., Chollet M., Song S., Lemke H., Liang M., Sokaras D., Alonso-Mori R., Zouni A., Messinger J., Bergmann U., Boal A. K., Bollinger J. M., Krebs C., Högbom M., Phillips G. N., Vierstra R. D., Sauter N. K., Orville A. M., Kern J., Yachandra V. K., Yano J. Nat. Methods . 2017;14:443. doi: 10.1038/nmeth.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Blaj G., Caragiulo P., Carini G., Carron S., Dragone A., Freytag D., Haller G., Hart P., Herbst R., Herrmann S., Hasi J., Kenney C., Markovic B., Nishimura K., Osier S., Pines J., Segal J., Tomada A., Weaver M. Synchrotron Radiat. News . 2014;27:14. doi: 10.1080/08940886.2014.930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chollet M., Alonso-Mori R., Cammarata M., Damiani D., Defever J., Delor J. T., Feng Y., Glownia J. M., Langton J. B., Nelson S., Ramsey K., Robert A., Sikorski M., Song S., Stefanescu D., Srinivasan V., Zhu D., Lemke H. T., Fritz D. M. J. Synchrotron Radiat. . 2015;22:503. doi: 10.1107/s1600577515005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Glöckner C., Kern J., Broser M., Zouni A., Yachandra V., Yano J. J. Biol. Chem. . 2013;288:22607. doi: 10.1074/jbc.m113.476622. [DOI] [PMC free article] [PubMed] [Google Scholar]