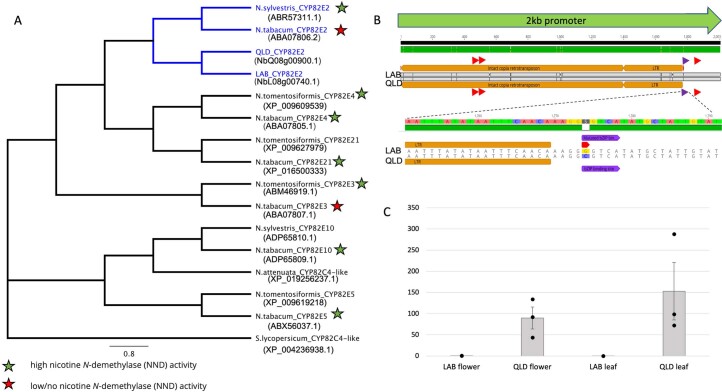
Extended Data Fig. 3. Cladogram of relationships of the nicotine demethylase genes in S. lycopersicum N. sylvestris, N. tabacum, N. tomentosiformis, N. attenuata, and N. benthamiana (LAB and QLD).
Cladogram of relationships of the nicotine demethylase genes in S. lycopersicum, N. sylvestris, N. tabacum, N. tomentosiformis, N. attenuata, and N. benthamiana (LAB and QLD). The highlighted clade contains the N. benthamiana CYP82E2 gene. Genes without stars represent proteins of uncharacterized nicotine N-demethylase activity. (B) Location of bZIP transcription factor binding motifs (red and purple triangles) in LAB and QLD 2kb promoter. The bottom panel shows the transversion in the third TF binding motif (purple triangles) that probably inhibits TF binding and expression of CYP82E2 in LAB. (C) Gene expression (TPM) of CYP82E2 in leaf and flower tissues of LAB and QLD. Error bars represent the standard error of the mean (n=3 biologically independent flower and leaf samples of LAB and QLD).