Figure 1. HMCES‐DNA‐protein crosslinks are reversible.
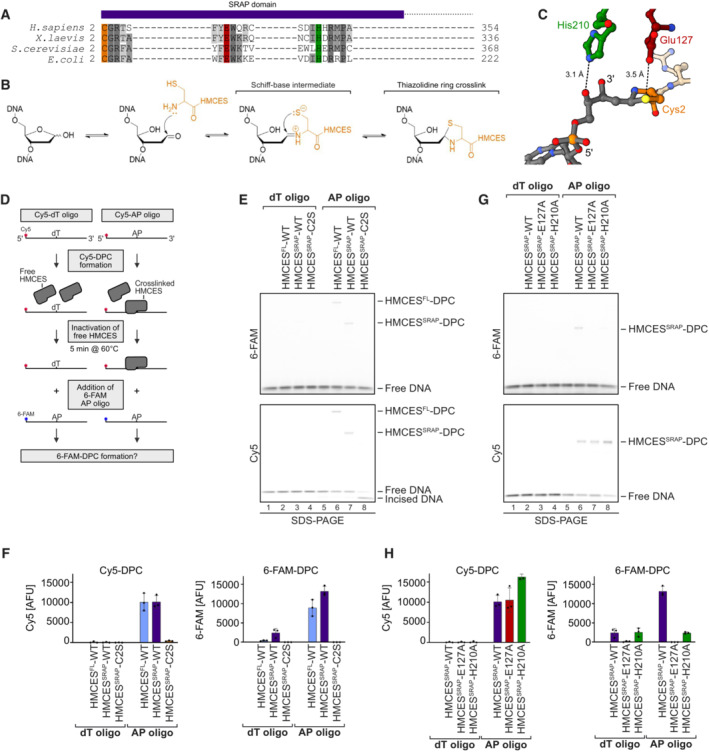
- SRAP domain sequence alignment highlighting key active site residues in H. sapiens, X. laevis, S. cerevisiae and E. coli HMCES homologues (Cys2 = orange, Glu127 = red and His210 = green).
- Proposed reaction mechanism of SRAP domain crosslinking to an AP site.
- Crystal structure of HMCES' active site crosslinked to an AP site. PDB: 6OE7 (Halabelian et al, 2019). DNA is shown in grey. Active site residues are coloured as in (A). Interatomic distances (Å) are labelled.
- Schematic of the assay shown in (E) and (G). HMCESFL and HMCESSRAP (WT or active site variants) were incubated for 1 h at 37°C with a Cy5‐labelled 30mer oligonucleotide containing either a dT or an AP site at position 15. Afterwards, non‐crosslinked HMCES was inactivated by heat denaturation at 60°C for 5 min. A second 6‐FAM‐labelled 30mer oligonucleotide containing an AP site was added and formation of 6‐FAM DPCs was assessed after an additional incubation for 120 min.
- HMCESFL‐ and HMCESSRAP‐WT and HMCESSRAP‐C2S‐DPC formation with Cy5‐ and 6‐FAM‐oligonucleotides was analysed using denaturing SDS–PAGE. Incised DNA is caused by spontaneous hydrolysis of the AP site.
- Quantification of DPC formation assays shown in (E), left panel: DPC formation to Cy5 oligonucleotide, right panel: DPC formation to 6‐FAM oligonucleotide.
- DPC formation of HMCESSRAP‐WT and variants (E127A or H210A) with Cy5‐ and 6‐FAM‐oligonucleotides was analysed using denaturing SDS–PAGE. Incised DNA is caused by spontaneous hydrolysis of the AP site.
- Quantification of DPC formation assays shown in (G), left panel: DPC formation to Cy5 oligonucleotide, right panel: DPC formation to 6‐FAM oligonucleotide.
Data information: Bar graphs in (F) and (H) show the mean of three independent experiments ± SD. Two WT data points are common between (F) and (H).
Source data are available online for this figure.
