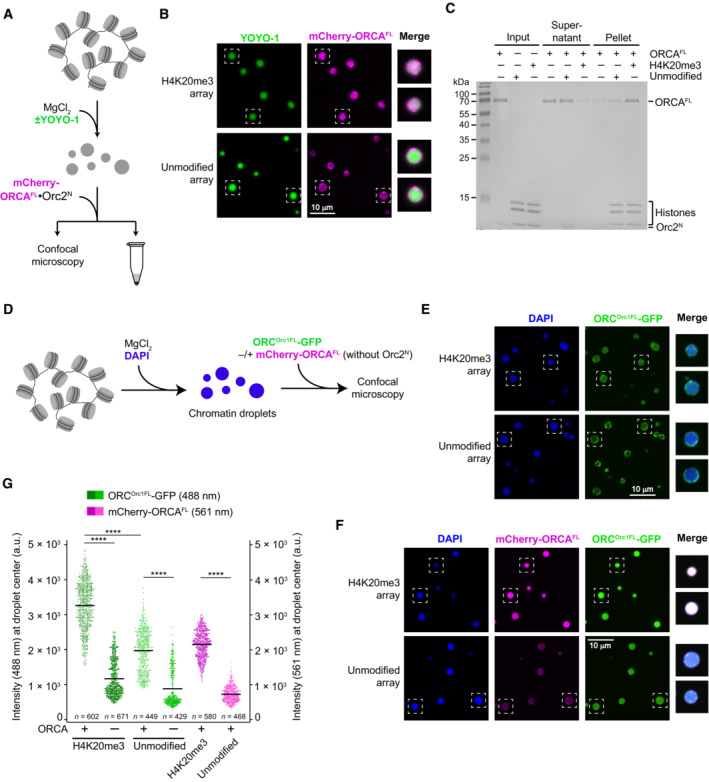
Figure EV4. ORCA and ORC are recruited to nucleosome arrays in a histone modification‐specific manner.
-
A–CORCA is recruited into preformed H4K20me3‐chromatin but not unmodified chromatin condensates. (A) Experimental setup. The endpoint is either confocal imaging of chromatin droplets or pelleting of nucleosome arrays by centrifugation. (B) Confocal microscopy images of unmodified and H4K20me3‐modified chromatin droplets after addition of mCherry‐labeled ORCA. Merged images for YOYO‐1 and mCherry channels are shown for boxed droplets. (C) Coomassie‐stained SDS–PAGE gel of histones and ORCA partitioning into supernatants and pellet fractions in nucleosome array pelleting assays. The difference between A‐C in this figure and Fig 6A–D is that here nucleosome arrays were first allowed to oligomerize prior to addition of ORCA.
-
D–GORCA enriches full‐length ORC in condensed nucleosome arrays in a histone modification‐dependent manner. (D) Schematic of experimental setup. ORCA and ORC are added to preformed chromatin droplets. (E) Confocal microscopy images of chromatin droplets with GFP‐labeled full‐length ORC in the absence of mCherry‐labeled ORCA. (F) Confocal microscopy images of chromatin droplets with GFP‐labeled full‐length ORC in the presence of mCherry‐labeled ORCA. (G) Quantification of ORC‐GFP and mCherry‐ORCA intensities at droplet centers. Statistical significance according to the Kruskal–Wallis test and Dunn's multiple comparisons test is indicated by asterisks (****P < 0.0001). The higher background recruitment of ORC to unmodified arrays as compared to Fig 6H is likely caused by interactions of the Orc1‐IDR with nucleosome array DNA.
