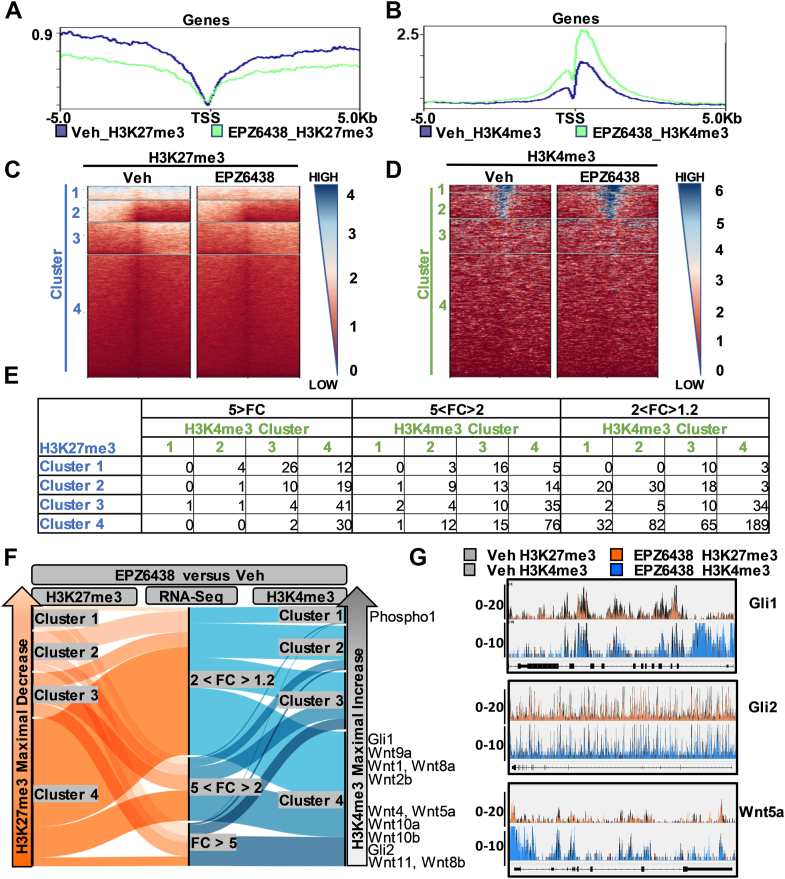
Figure 3.
WNT and hedgehog pathway genes associated with H3K27me3 reduction and H3K4me3 enrichment upon EPZ6438 treatment. H3K27me3 and H3K4me3 distribution at promoters from MC3T3 cells treated with Veh or EPZ6438 for 3 days. ChIP-seq analysis was performed within a region spanning ±5 kb around TSS. TSS profile plot of the H3K27me3 (A) and H3K4me3 (B) show the average ChIP signal for Veh (blue) and EPZ6438 (green) treatment. Profile heatmap around TSS of RefSeq genes was represented with gradient blue-to-red, this color indicates high-to-low counts in the corresponding region. Consensus H3K27me3 and H3K4me3 bounds peaks are separated into four clusters by K-means clustering (C–E). The changes in H3K27me3 (C, blue color) and in K3K4me3 (D, green color) are illustrated by heatmaps. Overlay between gene expression changes by RNA-seq analysis and alterations in H3K27me3 and H3K4me3 levels upon Ezh2 inhibition by EPZ6438 (E). Upregulated genes (RNA-seq) were subdivided into three groups based on fold change (FC) relative to vehicle (5 > FC, 5 < FC > 2, 2 < FC > 1.2). The changes in gene induction are correlated with alterations in H3K27me3 (blue color) and H3K4me3 (green color). The correlation between gene expression (RNA-seq) and H3K27me3 and H3K4me3 levels due to EPZ6438 treatment is further highlighted by an Alluvial plot (F). Genes associated with WNT and hedgehog pathways highlighted on the right. Genome browser view of H3K27me3 and H3K4me3 was performed for Gli1, Gli2, and WNT5a (G). ChIP, chromatin immunoprecipitation; TSS, transcriptional start site.