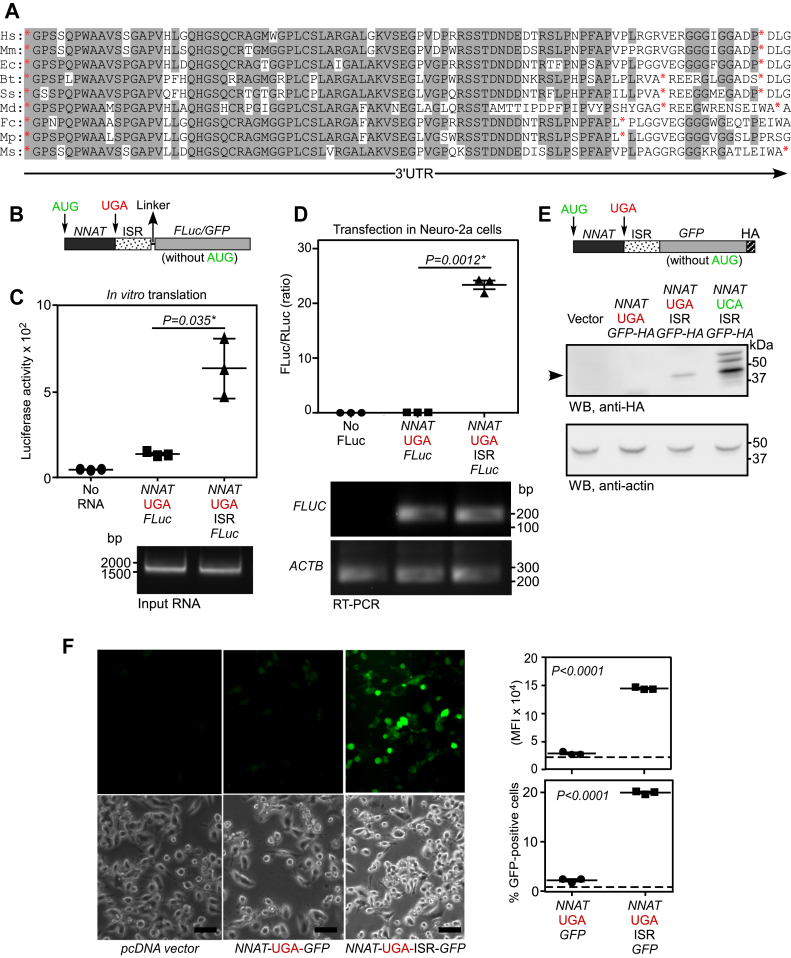
Figure 1.
NNAT mRNA exhibits termination codon readthrough.A, alignment of amino acid sequences predicted from the proximal 3′ UTR of NNAT mRNA belonging to multiple mammalian species. Positions of stop codons are indicated in red asterisks. Conserved residues are shown in gray background. B, schematic of the construct used in the luminescence- and fluorescence-based assays. C and D, results of luminescence-based TCR assay performed in vitro using rabbit reticulocyte lysate (C) and by transfection in Neuro-2a cells (D). Input RNA and firefly luciferase mRNA levels by RT-PCR are shown below the graphs. E, detection of TCR product by Western blot. Cells were transfected with indicated plasmids (see schematic above) and Western blotting was performed 48 h after that. Arrowhead indicates the TCR product. The result is a representative of two independent experiments. F, results of fluorescence-based TCR assay performed in Neuro-2a cells. Images of Neuro-2a cells transfected with indicated plasmids taken in a fluorescence microscope are shown. The scale bar represents 50 μm. Quantification of mean fluorescence intensity (MFI) and % of GFP-positive cells using flow cytometry are also shown. Dotted lines indicate the values for vector-transfected cells. All graphs (mean ± SD) are representatives of at least three independent experiments performed with triplicate samples. Statistical significance was calculated using two-tailed Student’s t test (∗, Welch’s correction was applied). Hs, Homo sapiens; Mm, Macaca mulatta; Ec, Equus caballus; Bt, Bos taurus; Ss, Sus scrofa; Md, Myotis davidii; Fc, Felis catus; Mp, Mustela putorius furo. Ms, Mus musculus; TCR, termination codon readthrough.