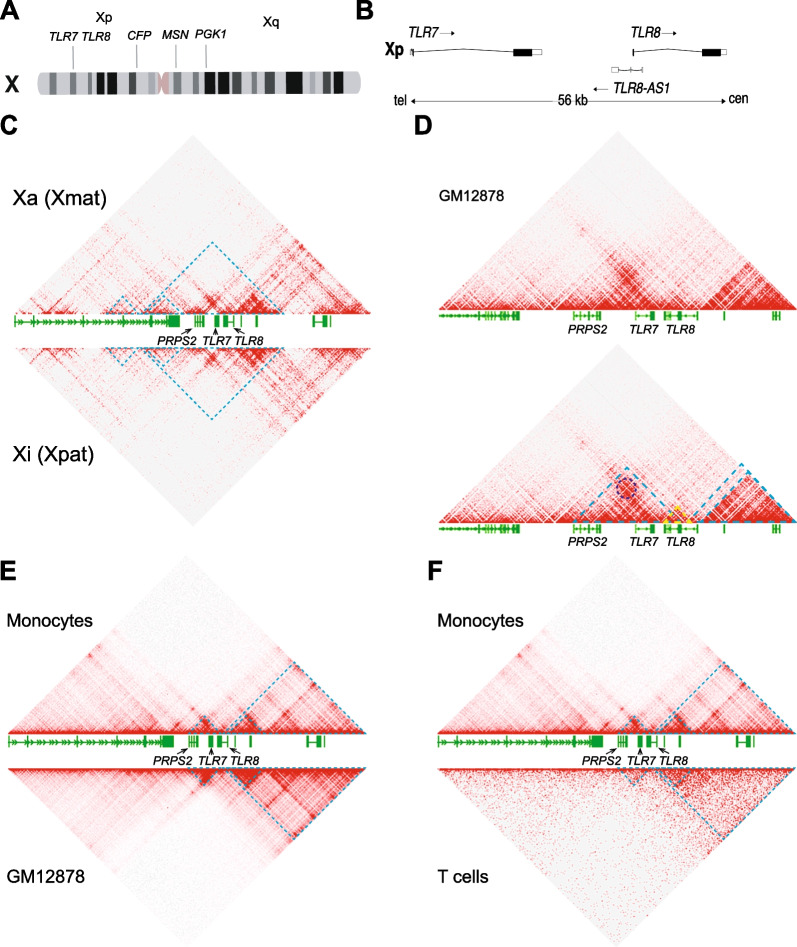
Fig. 1.
3D conformation of the TLR7–TLR8 genomic region in male and female cells. A Idiogram of the human X chromosome with the positions of the adjacent genes, TLR7 and TLR8, and three marker genes, CFP, MSN and PGK1, that do not evade X chromosome inactivation. B Map of the TLR7, TLR8-AS1 and TLR8 locus on Xp22.2; tel, cen denote the telomeric and centromeric ends of the map. C Hi-C map of interactions around the TLR7–TLR8 region on the active (Xa, top panel) or inactive (Xi, bottom panel) X chromosome at 5-kb resolution in GM12878 human female cells. D Hi-C map of interactions around the TLR7–TLR8 region at 1-kb resolution in GM12878 human female cells (datasets in C and D from Rao et al. [65]). The dark blue circle denotes a peak of interaction (loop anchor) between TLR7 and PRPS2 as determined by the Peak tool in the Juicebox software. Blue and yellow dashed lines represent domains of preferential interactions as defined by the Contact Domain tool in Juicebox. E, F Hi-C maps of interactions around the TLR7–TLR8 region at 5-kb resolution in human male monocytes E, F, top; dataset from Phanstiel et al., [66]) versus GM12878 human female cells (E, bottom, dataset from Rao et al., [65]) or male T cells (F, bottom panel, dataset from Zhang et al., [67]). Blue dashed lines represent domains of preferential interactions as defined by the Contact Domain tool in Juicebox