Fig. 6: T cells from patients carrying PSMC3/Rpt5 variants exhibit signs of protein homeostasis perturbations and alterations of the UPR, ISR and autophagy/mitophagy pathways.
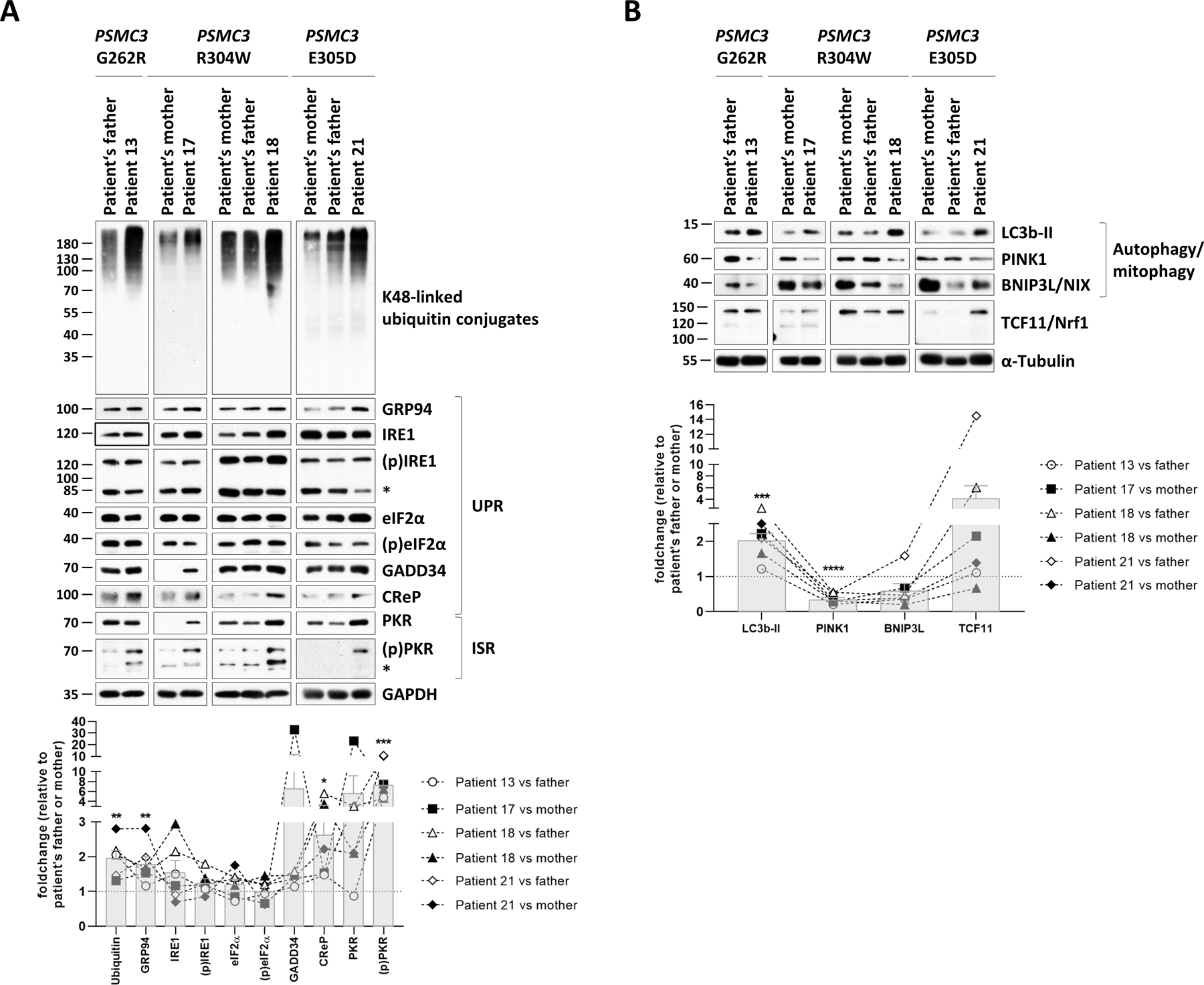
A. Top: five to twenty micrograms of RIPA lysates from T cells isolated from Patients #13, #17, #18 and #21 as well as related controls (PSMC3 index case’s father and/or mother) were separated by SDS-PAGE followed by Western-blotting using antibodies directed against K48-linked ubiquitin-modified proteins, GRP94, IRE1, phospho-IRE1, eIF2α, phospho-eIF2α, GADD34, CReP, PKR, phospho-PKR and GAPDH (loading control), as indicated. Bottom: Shown is the quantification of the Western-blots by densitometry. Data are presented as foldchanges in Patients #13, #17, #18 and #21 vs their father and/or mother whose densitometric measurements were set to 1 (gridline), as indicated. Columns indicate the foldchange mean values ± SEM calculated from the six normalizations. Statistical significance was assessed by unpaired Student’s test (*p<0.05, **p<0.01 and ***p<0.001). B. Top: RIPA-cell lysates from Patients #13, #17, #18 and #21 as well as their related controls (index case’s father and/or mother) were subjected to SDS-PAGE/Western-blotting using antibodies specific for LC3b, PINK1, BNIP3L and α-tubulin (loading control), as indicated. Bottom: quantification of the Western-blots by densitometry. Data are presented as protein foldchanges in Patients #13, #17, #18 and #21 vs their father and/or mother whose densitometric measurements were set to 1 (gridline), as indicated. Columns indicate the foldchange mean values ± SEM of the six normalizations. Statistical significance was assessed by unpaired Student’s test (***p<0.001 and ****p<0.0001).
