ABSTRACT
Epstein-Barr virus (EBV) is a human oncogenic γ-herpesvirus that establishes persistent infection in more than 90% of the world’s population. EBV has two life cycles, latency and lytic replication. Reactivation of EBV from latency to the lytic cycle is initiated and controlled by two viral immediate-early transcription factors, Zta and Rta, encoded by BZLF1 and BRLF1, respectively. In this study, we found that IQGAP2 expression was elevated in EBV-infected B cells and identified Rta as a viral gene responsible for the IQGAP2 upregulation in both B cells and nasopharyngeal carcinoma cell lines. Mechanistically, we showed that Rta increases IQGAP2 expression through direct binding to the Rta-responsive element in the IQGAP2 promoter. We also demonstrated the direct interaction between Rta and IQGAP2 as well as their colocalization in the nucleus. Functionally, we showed that the induced IQGAP2 is required for the Rta-mediated Rta promoter activation in the EBV lytic cycle progression and may influence lymphoblastoid cell line clumping morphology through regulating E-cadherin expression.
IMPORTANCE
Elevated levels of antibodies against EBV lytic proteins and increased EBV DNA copy numbers in the sera have been reported in patients suffering from Burkitt’s lymphoma, Hodgkin’s lymphoma, and nasopharyngeal carcinoma, indicating that EBV lytic cycle progression may play an important role in the pathogenesis of EBV-associated diseases and highlighting the need for a more complete mechanistic understanding of the EBV lytic cycle. Rta acts as an essential transcriptional activator to induce lytic gene expression and thus trigger EBV reactivation. In this study, scaffolding protein IQGAP2 was found to be upregulated prominently following EBV infection via the direct binding of Rta to the RRE in the IQGAP2 promoter but not in response to other biological stimuli. Importantly, IQGAP2 was demonstrated to interact with Rta and promote the EBV lytic cycle progression. Suppression of IQGAP2 was also found to decrease E-cadherin expression and affect the clumping morphology of lymphoblastoid cell lines.
KEYWORDS: Epstein-Barr virus, Rta, IQGAP2, lytic cycle
INTRODUCTION
Epstein-Barr virus (EBV) is a human oncogenic γ-herpesvirus that establishes persistent infection in more than 90% of the world’s population (1). This viral infection is the primary cause of infectious mononucleosis and is associated with several lymphoproliferative diseases, such as Burkitt’s lymphoma, Hodgkin’s lymphoma, NK/T cell lymphoma, post-transplant lymphoproliferative disorder (PTLD), and EBV+ diffuse large B cell lymphoma (DLBCL) (2). Although EBV remains predominantly in an asymptomatic latent state in immunocompetent individuals, impairment of the host immune system can stimulate EBV reactivation (3). EBV reactivation has been indicated to be triggered by several different conditions, such as stress, immunosuppression, and co-infection with various types of viruses (3, 4). Correlations between EBV reactivation and the pathogenesis of several diseases have been reported (4). One such example is PTLD, a serious complication involving dysregulated B cell lymphoproliferation that occurs in immunosuppressed individuals following organ transplantation (5). More recently, the effect of EBV reactivation in the setting of COVID-19 has also been examined. EBV reactivation was found to be associated with a longer ICU length of stay, although it was uncertain whether it would further affect mortality (6).
Reactivation of EBV from latency to the lytic cycle is initiated and controlled by two viral immediate-early (IE) transcription factors, Zta and Rta, encoded by BZLF1 and BRLF1, respectively. EBV lytic replication consists of a sequential expression of genes resulting in the production of viral particles and lysis of infected cells (7). A temporal cascade of three classes of genes is involved in the EBV lytic cycle, including IE, early, and late genes (8). Zta and Rta are initially activated by cellular transcription factors and further cooperate to induce the replication of viral DNA and the transcription of downstream early and late genes (9, 10). Both Zta and Rta have the ability to autostimulate their own expression, upregulate each other, and interact with or induce viral and host proteins (9, 10). These two proteins are both required to trigger and complete the viral lytic replication.
As mentioned above, Rta is an IE protein that acts as an important transcriptional activator to induce the expression of lytic genes and thus trigger EBV reactivation. Rta contains a DNA binding domain and a dimerization domain located in the N-terminus, while the transcriptional activation domain is located in the C-terminus (11). To activate gene expression and switch from latency to the lytic cycle, Rta exerts its transactivation function mainly in two ways: either via direct binding to specific sequences in the promoters of target genes or via cooperating with other factors to regulate gene expression. In the former way, Rta can selectively bind to an Rta-responsive element (RRE), a specific region with a GC-rich DNA sequence, in the promoter. For instance, Rta can directly bind to the promoters of some viral genes, such as BLLF1, BMRF1, and BARF1, as well as cellular genes like decoy receptor 3 (DcR3) (12 - 14). As for cases where the RRE is absent, Rta can alter gene expression through interaction with auxiliary cellular factors. As an example of the latter way, Rta can associate with Sp1 and methyl-CpG binding domain 1 (MBD1)-containing chromatin-associated factor 1 (MCAF1) to form an Sp1-MCAF-Rta complex that contributes to the autostimulation of Rp (15, 16). In this study, we discovered that IQGAP2, another protein involved in the regulation of diverse cellular activities, is upregulated via the direct binding of Rta to the RRE in the IQGAP2 promoter and also interacts with Rta to affect cellular functions and Rp activation.
The IQGAP family is a group of evolutionarily conserved scaffolding proteins, including IQGAP1, IQGAP2, and IQGAP3 (17). IQGAP family proteins interact with binding partners to mediate various cellular processes, such as cytokinesis, cell proliferation, cell migration, intracellular signaling, and cytoskeletal dynamics (17). The three IQGAP family proteins possess the same domains, including the calponin homology domain, the tryptophan-containing protein (WW) domain, the isoleucine-glutamine domain, the GAP-related domain responsible for the interaction with Cdc42 and Rac1, and the RasGAP C-terminus (RGCT) domain responsible for the binding of E-cadherin and β-catenin (17, 18). Though they comprise similar domains, the IQGAPs serve diverse roles. While IQGAP2 is 62% identical to IQGAP1, its biological functions are different from IQGAP1. In contrast to IQGAP1 and IQGAP3, decreased expression of IQGAP2 has been found to be associated with several human malignancies, suggesting that IQGAP2 functions as a tumor suppressor while IQGAP1 and IQGAP3 are oncoproteins. For instance, IQGAP1 and IQGAP3 are upregulated, while IQGAP2 is downregulated in hepatocellular carcinoma, for which patient prognosis has been found to be highly related to the expression of IQGAPs (19). In addition, downregulation of IQGAP2 has been shown to be associated with prostate cancer recurrence and metastasis (20). Downregulated IQGAP2 expression also promotes EMT and inhibits apoptosis as well as angiogenesis in breast cancer (21). In gastric cancer, IQGAP2 directly interacts with SHIP2 to elevate SHIP2 phosphatase activity, thus inhibiting the migration and invasion of gastric cancer cells (22). As scaffolding proteins, IQGAPs are also involved in viral infection processes, especially in viral maturation. IQGAP1 has been reported to facilitate the virulence of viruses from different families, such as Ebola virus (23), classical swine fever virus (24), and Moloney murine leukemia virus (25), while IQGAP2 has been reported to be required for IFN response against HCV infection and to influence HBV gene expression in hepatocytes (26).
In this study, we sought to determine the role of IQGAP2 in EBV infection and life cycle regulation. We found that IQGAP2 expression was elevated in EBV-infected B cells and identified Rta as a viral gene responsible for the IQGAP2 upregulation in both B cells and nasopharyngeal carcinoma (NPC) cell lines. Thus, we investigated the regulatory mechanism of Rta-mediated IQGAP2 induction in EBV-mediated immortalization of B lymphocytes and elucidated its biological significance in EBV-infected cells as well as its impact on EBV-associated disorders.
RESULTS
EBV infection and reactivation increase IQGAP2 expression
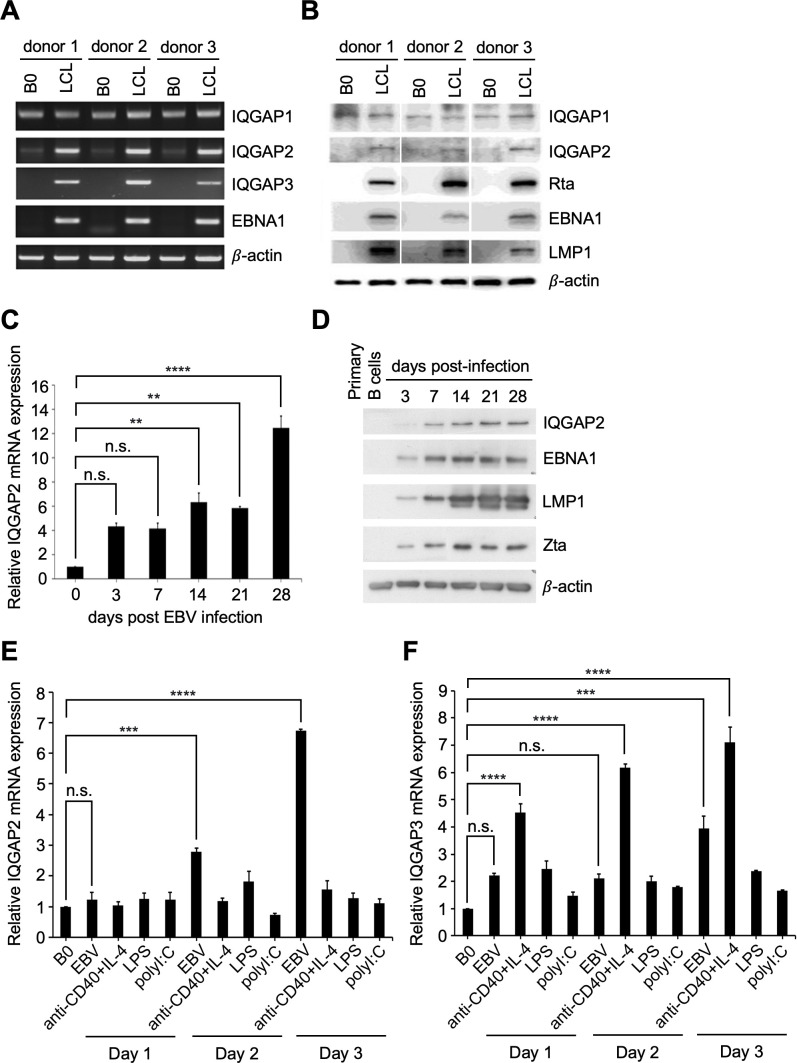
In this study, we examined the expression of the IQGAPs during EBV infection. Compared to uninfected B cells, the upregulation of IQGAP2 and IQGAP3 mRNA expression was observed in EBV-immortalized lymphoblastoid cell lines (LCLs) derived from three donors. In contrast, IQGAP1 mRNA was ubiquitously expressed in all tested donors, and no significant difference in expression was found between uninfected B cells and LCLs (Fig. 1A). Increased IQGAP2 expression in LCLs was also demonstrated at the protein level. Again, there was no significant difference in IQGAP1 expression when comparing uninfected B cells with LCLs (Fig. 1B). To further validate the effect of EBV infection on IQGAP2 expression, human CD19+ B lymphocytes were purified from the whole blood of healthy donors and infected with EBV strain B95.8. RNA and protein were isolated from EBV-infected B cells on days 3, 7, 14, 21, and 28 post-infection. Compared to the IQGAP2 mRNA expression in uninfected B cells, IQGAP2 mRNA expression was shown to be increased after EBV infection (Fig. 1C). The increased expression of IQGAP2 protein post-infection was also confirmed (Fig. 1D). Upregulation of IQGAP2 expression continued to be sustained on the days post-EBV infection leading up to and upon LCL establishment. In vitro, B cells alter cellular gene expression in response to different stimuli, leading to their activation or proliferation (27). In order to elucidate the underlying mechanisms of IQGAP2 upregulation in B lymphocytes, purified CD19+ B cells were either infected with EBV strain B95.8, stimulated with anti-CD40 plus IL-4, or triggered by lipopolysaccharides (LPS) or poly(I:C). The biologicals mimic T cell-dependent or -independent B cell activation, respectively. IQGAP2 was found to be upregulated prominently only after EBV infection and not after treatments with other stimuli (Fig. 1E). On the other hand, IQGAP3 expression was not only induced by EBV infection but also by other stimuli, such as anti-CD40 plus IL-4 (Fig. 1F). As a result, we focused on exploring the relationship between IQGAP2 and EBV infection in the follow-up experiments.
Fig 1.
IQGAP2 is induced by EBV infection and reactivation. (A and B) RNA and protein were harvested from paired uninfected primary B cells and EBV-immortalized LCLs. (A) mRNA expression of IQGAP1, IQGAP2, and IQGAP3 was detected by RT-qPCR. (B) Protein expression of IQGAP1, IQGAP2, Rta, EBNA1, LMP1, and β-actin was analyzed by western blotting. (C and D) Primary B cells were infected with EBV strain B95.8, and cellular RNA was harvested on the indicated days post-infection. (C) IQGAP2 mRNA expression was measured by RT-qPCR and normalized to β-actin expression. Fold changes were acquired by comparing normalized expression levels of IQGAP2 in EBV-infected B cells to those in uninfected primary B cells. (D) IQGAP2 protein expression was analyzed by western blotting. LMP1 and EBNA1 served as the EBV infection markers, Zta served as the lytic progression marker, and β-actin served as the internal loading control. (E and F) Purified primary B cells were infected with EBV, stimulated by anti-CD40 + IL-4, or triggered by LPS or poly(I:C). Cellular RNA was harvested on days 1, 2, and 3 post-treatment and measured by RT-qPCR. Expression of (E) IQGAP2 and (F) IQGAP3 was normalized to the expression of β-actin. Fold changes were acquired by comparing normalized expression levels in treated B cells to those in untreated B cells. **** indicates P < 0.0001, *** indicates P < 0.001, and ** indicates P < 0.01; n.s. stands for not significant.
Rta is responsible for IQGAP2 upregulation
Having demonstrated the association between IQGAP2 upregulation and EBV infection, we sought next to determine its potential relevance to the EBV life cycle. EBV-negative (Akata−) and EBV-positive (Akata+) cells were treated with or without 0.5% goat anti-human immunoglobulin G to understand the effect of EBV reactivation on IQGAP2 expression. The results of RT-PCR and western blotting detected significantly higher levels of IQGAP2 mRNA and proteins when EBV entered the lytic cycle (Fig. 2A and B). In order to identify the viral gene responsible for the IQGAP2 upregulation, EBV-negative NPC TW01 cells were transfected with various important viral genes, including those for EBNA1-6, LMP1, LMP2A, EBER1/2, BARF0, Rta, Zta, and BGLF4. The RT-PCR results identified the lytic protein Rta as the only EBV viral product to induce IQGAP2 mRNA expression. On the other hand, none of the tested EBV viral products led to differences in IQGAP1 or IQGAP3 expression (Fig. 2C). The upregulation of IQGAP2 was also observed in TW01 cells and BJAB cells transfected with Rta-expressing plasmids (Fig. 2D). Together, these results suggest that Rta is required for the induction of IQGAP2.
Fig 2.
Rta is responsible for the induction of IQGAP2 expression. (A and B) EBV-negative (Akata−) and EBV-positive (Akata+) cells were treated without or with 0.5% goat anti-human IgG. (A) RNA and (B) protein were extracted for RT-PCR and western blotting, respectively. Expression of IQGAP2, EBNA1, Zta, and β-actin was detected. (C) Expression of IQGAPs was analyzed in TW01 cells transfected with the indicated EBV viral genes. mRNA expression was assessed by RT-PCR analysis 72 h post-transfection. (D) TW01 cells were transfected with pSG5 or pSG5-Rta using NTR II, and cell lysates were harvested 72 h post-transfection. Transient expression of Rta was achieved by electroporation in BJAB cells, and cell lysates were prepared 48 h later. The obtained proteins were analyzed by western blotting to detect the expression of IQGAP2 and Rta. β-actin served as the internal control.
DNA binding and transcriptional activation domains of Rta enhance IQGAP2 expression
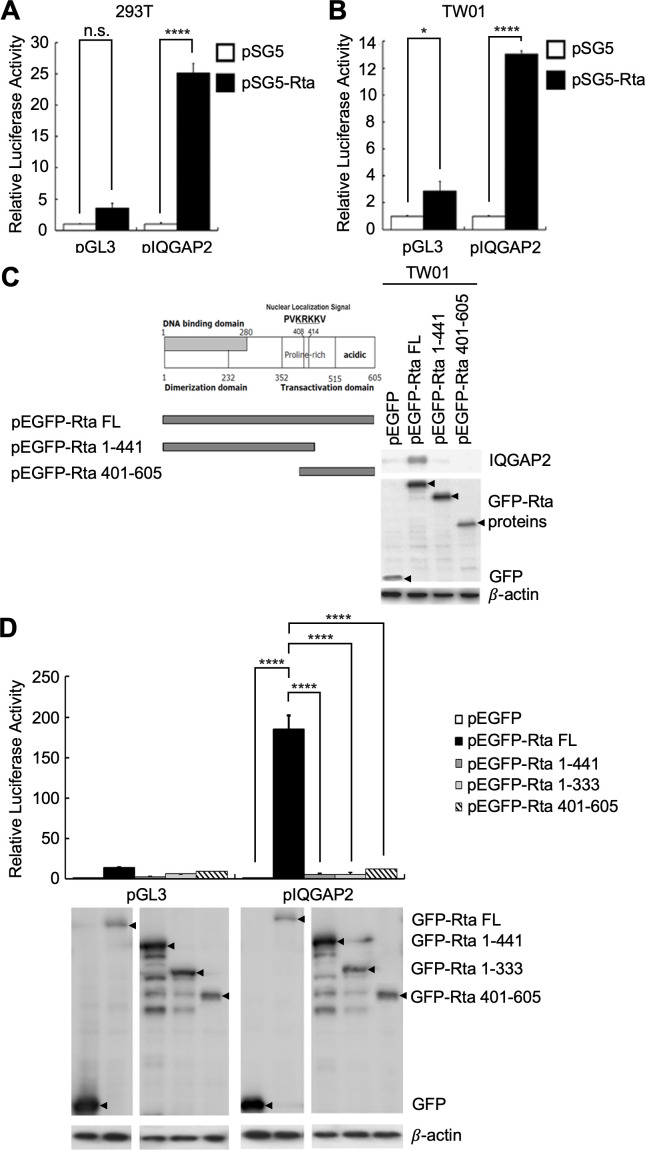
As a viral transactivator, Rta can transactivate many cellular and viral genes via directly binding to RRE-containing gene promoters or interacting with DNA-bound transcription factors to be targeted to binding sites in gene promoters (7, 14). To further understand how Rta mediates IQGAP2 upregulation, we investigated the transactivational activities of Rta at the IQGAP2 promoter. The results of reporter assays conducted with 293T cells and TW01 cells showed that Rta was able to activate the IQGAP2 promoter in both cell lines (Fig. 3A and B). As three major functional domains have been identified in the Rta protein, including a DNA binding domain that contains a dimerization domain located in the N-terminus as well as a transcriptional activation domain located in the C-terminus, we sought to identify the domains essential for transactivation. TW01 cells were transfected with truncated N- or C-terminal Rta-expressing plasmids to discern each domain’s contribution to the induction of IQGAP2. The nuclear localization signal (NLS) was retained on both truncated forms of Rta to enable these truncated proteins to enter the nucleus and function as transactivators. The results of western blotting demonstrated that the full length of Rta was required to induce IQGAP2 expression. Deletion of either the N- or C-terminus resulted in failure to induce IQGAP2 expression (Fig. 3C). A follow-up reporter assay was performed to further confirm the necessity of both the N- and C-termini for IQGAP2 promoter activation. A transcriptional activation domain-deleted Rta without NLS (GFP-Rta 1–333) was also used in this experiment. The results revealed that only full-length Rta can activate the IQGAP2 promoter (Fig. 3D). Altogether, the results indicate that Rta requires both the DNA binding/dimerization and transcriptional activation domains to induce IQGAP2 expression.
Fig 3.
DNA binding and transcriptional activation domains of Rta induce IQGAP2 expression. (A) 293T and (B) TW01 cells were transfected with pGL3 or pGL3-pIQGAP2, pSG5 or pSG5-Rta, and pEGFP-C1. The luciferase activity of each transfectant was normalized to its GFP fluorescence intensity and standardized using the normalized luciferase activity of the vector control cells. (C) A schematic of a full-length Rta containing a DNA binding domain, dimerization domain, and transcriptional activation domain (left panel). TW01 cells were transfected with pEGFP-C1, pEGFP-Rta (full-length Rta-expressing plasmid), pEGFP-Rta 1-441, or pEGFP-Rta 401-605 using NTR II. Cell lysates were harvested 72 h post-transfection. Expression of IQGAP2, GFP, full-length and truncated forms of GFP-Rta proteins, and β-actin was analyzed by western blotting (right panel). (D) 293T cells were transfected with pGL3 or pGL3-pIQGAP2, and pEGFP-C1, pEGFP-Rta, pEGFP-Rta 1-441, pEGFP-Rta 1-333, or pEGFP-Rta 401-605. Luciferase activities were measured, and relative activities were calculated according to GFP fluorescence intensity (top panel). Transfected cells were harvested, and cell lysates were subjected to protein analysis to verify the expression of GFP, full-length and truncated forms of GFP-Rta proteins, and β-actin (bottom panel). **** indicates P < 0.0001, * indicates P < 0.05; n.s. stands for not significant.
Rta increases IQGAP2 expression through direct binding to the IQGAP2 promoter
Next, we sought to further clarify the underlying regulatory mechanism of Rta in IQGAP2 upregulation. Considering that Rta can directly and indirectly induce gene expression, several putative transcription factor binding sites in the IQGAP2 promoter were predicted using ALGGEN-PROMO software, including upstream stimulating factor (USF), E2 factor (E2F), specificity protein 1 (Sp1), activator protein 1 (AP1), and Rta-responsive element (Fig. 4A). Serial deletion constructs of the promoter were constructed for reporter assays. The results revealed the importance of the RRE site for the Rta-mediated IQGAP2 transactivation (Fig. 4B). Of note, since Sp1 is largely involved in Rta-mediated gene transactivation and several putative Sp1 binding sites were found in the IQGAP2 promoter, we used shSp1 to address the effect of Sp1 on Rta-mediated IQGAP2 expression. The results demonstrated that Sp1 may not be involved in Rta-mediated IQGAP2 transactivation (Fig. 4C). In order to confirm the direct binding of Rta to the IQGAP2 promoter, chromatin immunoprecipitation was performed with pEGFP-C1- and pEGFP-Rta-transfected 293T cells. GFP-Rta-binding DNA complexes were analyzed using primers targeting a region in the IQGAP2 promoter spanning the putative RRE. The results revealed that Rta is directly associated with the IQGAP2 promoter (Fig. 4D).
Fig 4.
Rta binds directly to the IQGAP2 promoter to increase IQGAP2 expression. (A) Rta-related DNA binding sites in the IQGAP2 promoter were predicted with ALGGEN-PROMO. The schematic illustrates the putative binding sites of the listed transcription factors in the IQGAP2 promoter, including USF, E2F, Sp1, AP1, and RRE. (B) 293T cells were transfected with pSG5 or pSG5-Rta, pGL3 or one of the serial 5′-deleted pGL3-pIQGAP2 luciferase reporter plasmids, and pEGFP-C1 as the transfection control. After 72 h, the luciferase activity of each transfectant was normalized to its GFP fluorescence intensity and standardized using the normalized luciferase activity of the vector control cells. (C) 293T cells were infected with shLuc- or shSp1-expressing lentivirus and underwent puromycin selection for 48 h. Control and Sp1-suppressed 293T cells were then transfected with pGL3 or pGL3-pIQGAP2, and pSG5 or pSG5-Rta. After 72 h, the luciferase activity of each transfectant was normalized to its GFP fluorescence intensity and standardized using the normalized luciferase activity of the vector control cells. (D) 293T cells were transfected with pEGFP-C1 or pEGFP-Rta for 72 h and lysed. Complexes of DNA and GFP-Rta were immunoprecipitated from the cell lysates using anti-GFP or mouse IgG antibody. The IQGAP2 promoter (pIQGAP2) DNA was detected in the immunocomplexes precipitated with the anti-GFP antibody by PCR analysis (top panel). The immunocomplexes were analyzed by western blotting to verify the protein expression of GFP or GFP-Rta in transfected cells (bottom panel). Protein extracts from pEGFP-C1- or pEGFP-Rta-transfected cells before performing immunoprecipitation were defined as input. **** indicates P < 0.0001 and * indicates P < 0.05; n.s. stands for not significant.
Rta colocalizes with IQGAP2 in the nucleus
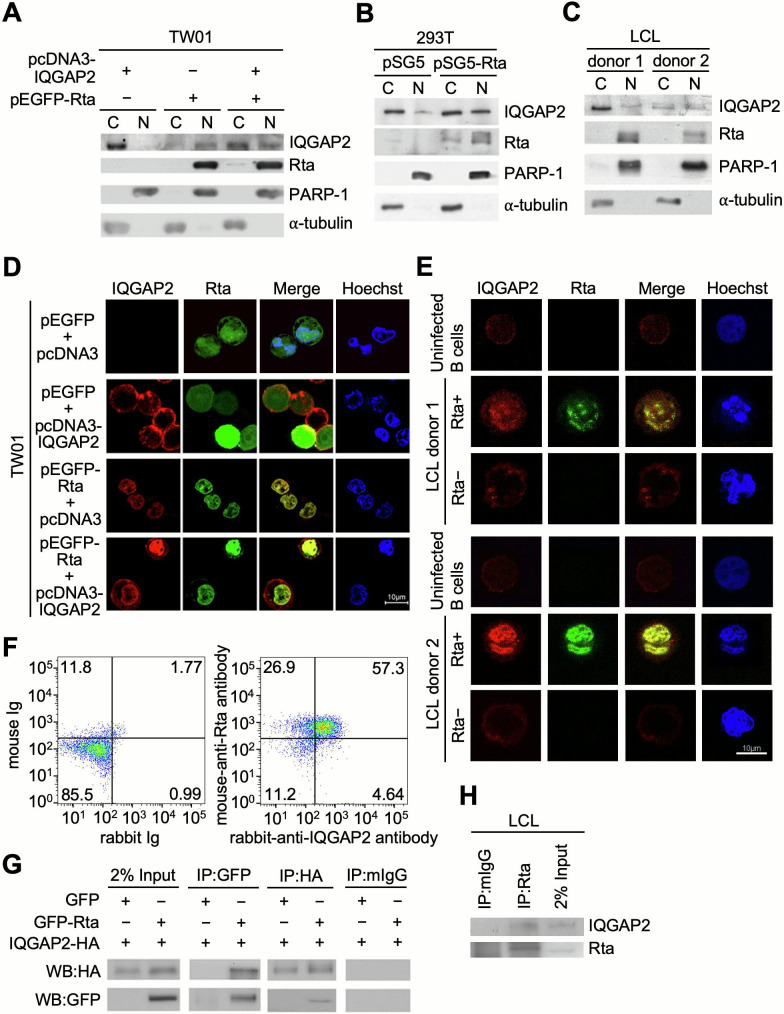
IQGAP2 is thought to interact with different proteins to regulate cellular functions (17, 18). In order to elucidate the biological roles of IQGAP2 in Rta induction, the cellular distribution of IQGAP2 was investigated. In TW01 cells, IQGAP2 was mainly found in the cytoplasm when only IQGAP2 was expressed, which is consistent with previous findings (17, 18). However, an increased percentage of IQGAP2 was found in the nucleus in the presence of Rta (Fig. 5A). We also found the same results in 293T cells transfected with pSG5 or pSG5-Rta (Fig. 5B). In LCLs from two different donors, IQGAP2 was found in the nuclear fraction as well as the cytoplasmic fraction when both Rta and IQGAP2 were expressed (Fig. 5C). Together, these results suggest that IQGAP2 is partially translocated from the cytoplasm into the nucleus when both Rta and IQGAP2 are expressed. Further experiments using confocal microscopy were also performed to locate the subcellular distribution of Rta and IQGAP2. In the absence of Rta, IQGAP2 was observed to be located in the cortex of TW01 cells (Fig. 5D). However, when IQGAP2 was induced by Rta or when Rta- and IQGAP2-expressing plasmids were co-transfected into TW01 cells, IQGAP2 was partially redistributed into the nucleus and colocalized with Rta (Fig. 5D). Similar results were observed in LCLs (Fig. 5E). Approximately 20% of LCLs undergo spontaneous lytic cycle progression, while the rest remain in latency at any given time under normal conditions (28), resulting in the presence of both Rta-negative and Rta-positive LCLs. The basal expression of IQGAP2 could be detected and found to be aggregated near the plasma membrane region in uninfected B cells and Rta-negative LCLs. However, in Rta-positive LCLs, IQGAP2 seems to colocalize in the nucleus with Rta (Fig. 5E). These results suggest that Rta may recruit IQGAP2 to the nuclear region of the cell. Flow cytometry results also demonstrated that 57.3% of the LCLs cultured in Roswell Park Memorial Institute medium (RPMI) supplemented with low fetal bovine serum (FBS) percentage for 5 d to increase lytic cycle antigen expression were Rta+IQGAP2+ double positive (Fig. 5F). In addition, co-immunoprecipitation (co-IP) results showed the interaction between co-expressed IQGAP2 and Rta proteins in transfected 293T cells (Fig. 5G). Co-IP using LCLs cultured in RPMI supplemented with low FBS percentage for 5 d further confirmed the endogenous association of IQGAP2 with Rta (Fig. 5H). In sum, the results together suggest that Rta may interact with IQGAP2 and colocalize in the nucleus.
Fig 5.
IQGAP2 interacts and is colocalized with Rta in the nucleus. (A) TW01 cells were transfected with pEGFP-C1 or pEGFP-Rta, and pcDNA3 or pcDNA3-IQGAP2-HA. At 72 h post-transfection, cells were harvested for subcellular fractionation and then analyzed by western blotting for detection of IQGAP2 and Rta expression in the cytoplasm (C) and nucleus (N). (B) 293T cells were transfected with pSG5 or pSG5-Rta and subjected to subcellular fractionation at 72 h post-transfection. Expression of IQGAP2 and Rta in the cytoplasm and nucleus was analyzed by western blotting. (C) The cytoplasm and nucleus of LCLs were separated and analyzed by western blotting. Poly (ADP-ribose) polymerase-1 (PARP-1) and α-tubulin were used as nuclear and cytoplasmic markers, respectively. (D) TW01 cells were seeded onto coverslips and transfected with pcDNA3 or pcDNA3-IQGAP2-HA, and pEGFP-C1 or pEGFP-Rta. After 72 h, cells were fixed with 2% formaldehyde and permeabilized with 0.1% Triton X-100. (E) Human CD19+ uninfected B lymphocytes and EBV-immortalized LCLs were fixed with 2% formaldehyde and permeabilized with 0.1% Triton X-100. Confocal microscopy was utilized to observe the relative expression of IQGAP2 (red) and Rta (green) in the cells. Hoechst 33342 (blue) was used to indicate the nuclei of the cells. (F) LCLs were cultured in RPMI supplemented with 5% FBS for 5 d. Cells were harvested and incubated with irrelevant monoclonal mouse IgG and rabbit IgG, or mouse anti-Rta and rabbit anti-IQGAP2 antibodies. After 30 min, cells were washed with PBS and then cells were stained with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG and rhodamine-conjugated goat anti-rabbit IgG antibodies for 30 min. Rta+IQGAP2+ LCLs were analyzed by flow cytometry. (G) 293T cells were transfected with both pcDNA3-IQGAP2-HA and pEGFP-C1 or pEGFP-Rta. After 72 h, cells were harvested and pre-cleared with Protein A beads. Anti-GFP antibody was used to bind GFP-Rta, while an anti-HA tag antibody was used to bind IQGAP2-HA. Protein-antibody complexes were then precipitated with Protein A beads and analyzed by western blotting to demonstrate the interaction between IQGAP2 and Rta. (H) LCLs were cultured in RPMI supplemented with 5% FBS for 5 d. Cells were harvested and pre-cleared with irrelevant monoclonal antibodies. Anti-Rta antibody was used to bind Rta. Protein-antibody complexes were then precipitated with Protein A beads and analyzed by western blotting to demonstrate the interaction between IQGAP2 and Rta.
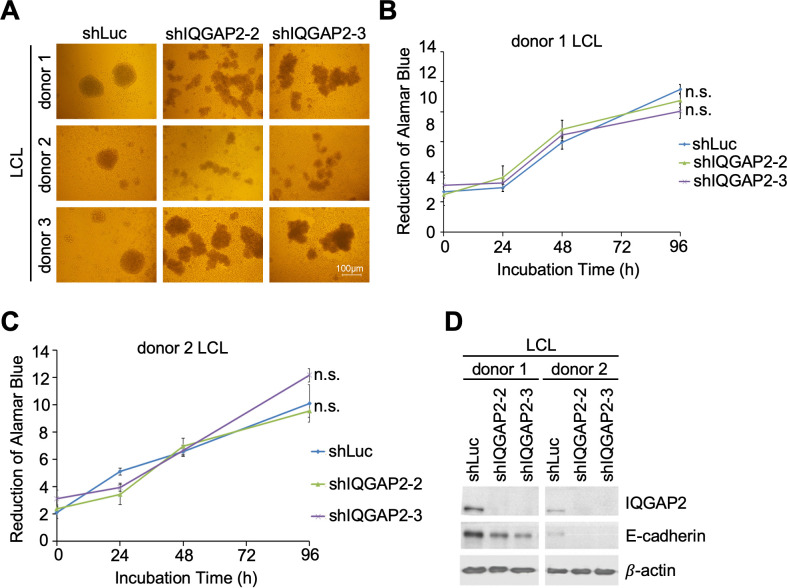
IQGAP2 knockdown lowers E-cadherin expression and alters the clumping morphology of LCLs
Finding the interaction between IQGAP2 and Rta as well as their colocalization in the nucleus, we next examined the biological effect of IQGAP2 on EBV-immortalized LCLs. First, we found that the clumping morphology of LCLs changed when IQGAP2 was knocked down in LCLs. The clumping of LCLs became scattered rather than spherical (Fig. 6A). Since this alteration of the clumping pattern could be the result of alteration in cell proliferation, alamarBlue assay was performed to determine the proliferation rate of LCLs in which IQGAP2 was knocked down. The results showed that the LCLs from two donors with suppressed IQGAP2 expression had a comparable proliferation rate to that of the negative control (Fig. 6B and C). We further inspected the possibility of altered cell-cell adhesion influencing the clumping of LCLs. A previous study has shown that IQGAP2 binds with E-cadherin through its RGCT domain (18). On account of the role adhesion molecule E-cadherin plays in cell-cell interaction, the expression of E-cadherin in LCLs was examined with western blotting. The results demonstrated that IQGAP2 knockdown led to lower E-cadherin expression in LCLs (Fig. 6D). These results suggest that it is possible that IQGAP2 may mediate cell-cell adhesion by playing an undetermined role in regulating E-cadherin expression and thus affect the clumping of LCLs.
Fig 6.
Knockdown of IQGAP2 alters LCL clumping morphology and decreases E-cadherin expression. (A–D) LCLs were infected with shLuc-, shIQGAP2-2-, or shIQGAP2-3-expressing lentivirus for 5 d and then underwent 48 h of puromycin selection. (A) Cells were then seeded in 96-well plates and incubated for 24 h. Images of LCL clumping morphology were taken under a bright-field microscope. LCLs from (B) donor 1 and (C) donor 2 were seeded in 96-well plates, and cell viability was measured with the alamarBlue assay. Cell proliferation rate was obtained by comparing the number of viable cells over time. n.s. stands for not significant. (D) Cells were then harvested and analyzed by western blotting to detect E-cadherin expression. Anti-IQGAP2 antibody was used to confirm knockdown efficacy. β-actin served as the internal control.
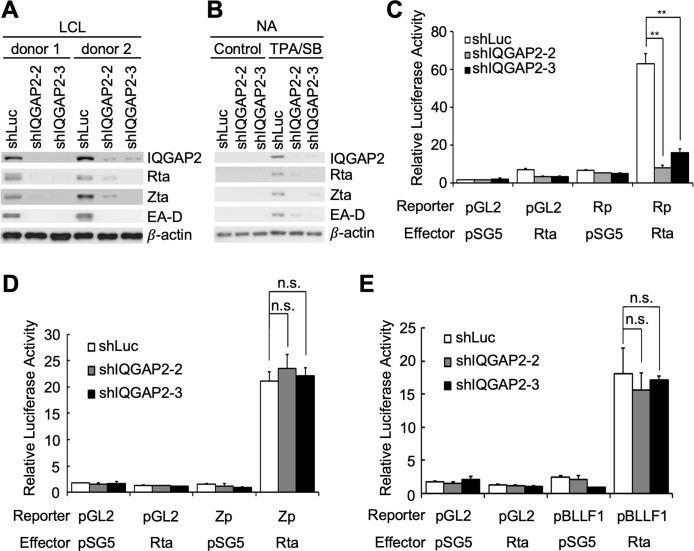
IQGAP2 is required for EBV lytic replication
Knowing that the functions of IQGAP2 are varied and highly related to its binding partner (18), we speculated that IQGAP2 may play a role in the initiation of EBV lytic replication by helping Rta to carry out viral biological activities. To test this speculation, LCLs were infected with lentiviruses containing shRNA-expressing plasmids designed to knockdown IQGAP2. It was observed that the suppression of IQGAP2 resulted in lower expression of lytic proteins, including Rta, Zta, and EA-D, suggesting the reduction of spontaneous lytic replication in LCLs (Fig. 7A). To check whether this observation would be found in other EBV-infected cells, an NPC cell line latently infected with EBV, NA, was also used. Following treatment of NA cells transfected with the aforementioned shRNA-expressing plasmids with 12-O-tetradecanoylphorbol-13-acetate (TPA) and sodium butyrate (SB), decreased expression of the same lytic proteins also indicated that knockdown of IQGAP2 impairs chemically induced lytic progression in NA cells (Fig. 7B). Reporter assays were further conducted to understand the influence of IQGAP2 on the activation of Rta-responsive promoters, including Rp, Zta promoter (Zp), and BLLF1 promoter (pBLLF1). The results showed that Rp could not be activated by Rta when IQGAP2 was knocked down in 293T cells, indicating the participation of IQGAP2 in Rp autoactivation (Fig. 7C). On the other hand, the results showed that IQGAP2 depletion did not affect Rta-mediated Zp or pBLLF1 activation (Fig. 7D and E).
Fig 7.
Knockdown of IQGAP2 affects EBV lytic progression. (A) LCLs were infected with shLuc-, shIQGAP-2–2-, or shIQGAP2-3-expressing lentivirus for 5 d, and (B) NA cells were transfected with shLuc-, shIQGAP-2–2-, or shIQGAP2-3-expressing plasmids using NTR II for 72 h. Two types of cells both underwent 48 h of puromycin selection. The selected NA cells were then treated with TPA and SB for 72 h. Cell lysates were harvested, and expression of Rta, Zta, and EA-D was observed to detect lytic progression. Anti-IQGAP2 antibody was used to confirm knockdown efficacy. β-actin served as the internal control. (C–E) 293T cells were transfected with shLuc-, shIQGAP2-2-, or shIQGAP2-3-expressing plasmids, and further transfected with (C) pGL2 or pGL2-Rp, pSG5 or pSG5-Rta, and pEGFP-C1, (D) pGL2 or pGL2-Zp, pSG5 or pSG5-Rta, and pEGFP-C1, or (E) pGL2 or pGL2-pBLLF1, pSG5 or pSG5-Rta, and pEGFP-C1 for reporter assays. The luciferase activity of each transfectant was normalized to its GFP fluorescence intensity, and the relative luciferase activity was obtained by setting the normalized luciferase activity of cells transfected with pGL2 and pSG5 as 1. ** indicates P < 0.01; n.s. stands for not significant.
DISCUSSION
EBV often manipulates cellular factors to benefit its own infection and pathogenesis; thus, abnormal expression of cellular factors in EBV infection may be a sign of viral regulation (12, 13). The IE transactivator Rta is critical for EBV lytic progression as it activates the transcription of lytic genes. Growing evidence also points to the importance of Rta in regulating cellular factors (12). Here, we demonstrated the novel finding that Rta also plays a crucial role in regulating cellular factor IQGAP2 via direct binding to the RRE in the IQGAP2 promoter and directly activating IQGAP2 transcription. The GC-rich RRE was first defined on EBV viral promoters (29), and the binding sequence of Rta on the EBV genome has been extensively studied throughout the years. However, the binding of Rta to cellular genes is seldom discussed. In 2007, Rta was shown to increase DcR3, a soluble decoy receptor that belongs to the tumor necrosis factor receptor superfamily, by direct binding to its promoter (13). In our study, in addition to the direct interaction between Rta and the IQGAP2 promoter, we discovered that the RRE located in the pIQGAP2 region -157/-140 is important to the Rta-induced IQGAP2 upregulation as deletion of this sequence resulted in failure to activate Rta-mediated IQGAP2 transcription.
We also demonstrated that IQGAP2 associates with Rta, and they are found to colocalize in the nucleus. Furthermore, we showed that IQGAP2 plays an important role in Rp activation in the presence of Rta. However, we did not find IQGAP2 to be involved in Rta-mediated Zp or pBLLF1 transactivation. The reason behind this phenomenon could be that Rta activates these promoters through different pathways. Rta activates pBLLF1 by direct binding to an RRE (29). On the contrary, Rta regulates Rp and Zp through indirect, non-Rta-DNA binding ways as they lack an RRE. It has been reported that Rta interacts with cellular factors for lytic progression. For example, Rta mediates Rp activation via interacting with Sp1 and Sp1-interacting protein MCAF1, forming an Sp1-MCAF1-Rta complex that enhances Sp1-mediated transcription (15). Meanwhile, the phosphorylated ATF2 transcription factor is required for Rta to activate Zp (9). In addition, Rta also transactivates the BALF5 promoter through two cellular transcription factors, USF and E2F (30), as well as interacts with cellular transcription factor Oct-1 to enhance transactivation of EBV lytic genes such as those for SM, EA-D, BHLF1, BHRF1, and Zta (31). In our lab, we also found previously that Rta interacts with TSG101 to activate the expression of EBV late genes, including those for BcLF1, BDLF3, BILF2, BLLF1, and BLRF2 (14).
In addition to activating the viral genes required for EBV lytic progression, Rta also influences cellular functions that contribute to the viral lytic cycle through various pathways. For example, Rta transactivates the c-myc promoter, an important human oncogene, via an indirect mechanism (32). As another example, Rta interacts with retinoblastoma protein (Rb), which acts as a tumor suppressor, to alleviate the Rb-mediated repression of E2F. The binding of Rta to Rb and the further correlation with E2F have been shown to facilitate the entry into G1/S in the cell cycle and viral DNA synthesis during EBV lytic replication (33, 34). Furthermore, cellular fatty acid synthase (FAS) activity is important for EBV replication, and Rta increases FAS expression through a stress mitogen-activated protein kinase-dependent pathway (35). Other cellular genes involved in the cell cycle that have been reported to be upregulated by Rta through Sp1 include tumor suppressor p16, cyclin-dependent kinase inhibitor p21, and small nuclear ribonucleoprotein polypeptide N (15).
Our work here demonstrated that IQGAP2 is required for EBV to enter the lytic cycle, and this prerequisite is correlated to the Rta-induced Rp activation. Considered together with the findings of the aforementioned studies, it is possible that IQGAP2, along with Rta, has the ability to alter cellular gene expression and affect cellular activities such as those involved in the cell cycle to promote EBV lytic progression.
In this study, we found that IQGAP2 and IQGAP3 expressions increase when purified human CD19+ B lymphocytes are immortalized into LCLs, while only IQGAP3 expression increases when these B cells are treated with biological stimuli other than EBV. IQGAP3 correlates with cell proliferation and cytokinesis and could be one of the cellular factors that EBV manipulates to promote B cell proliferation. However, from our data, the possibility that IQGAP3 upregulation results from B lymphocyte proliferation rather than being the cause of it could not be ruled out. Thus, while our findings successfully elucidated several roles that IQGAP2 may play in EBV infection, yet more effort is needed to fully understand the interaction between IQGAPs and EBV.
IQGAPs have also been reported to be involved in the modulation of the cytoskeleton and cell adhesion through binding to various effectors. Examination of the cellular distribution of IQGAPs has revealed that they are mostly found in the cytoplasm. Few studies have further focused on the subcellular localization of IQGAP2. In prostate cancer cell lines, LNCaP and DU145, IQGAP2 has been observed in the cytosol (36). In gastric cancer cell lines, including HSC-59, NUGC4, MKN45, and TGBC11TKB, IQGAP2 has been limited to the cell membrane (37). IQGAP2 has also been found in the filopodial extensions of activated platelets (38).
Although the effects of IQGAP1 and IQGAP2 have been found mainly to be cytoplasmic, several studies have indicated that they may be nuclear as well. IQGAP1 interacts with several nuclear proteins, including estrogen receptor (ER) α, ERβ, nuclear factor of activated T cells 1, and nuclear factor-erythroid-related factor 2 (Nrf2), and has been shown to serve as a coregulator with ERα or Nrf2 to enhance estradiol-induced transcriptional activation or activation of the MEK-ERK-Nrf2 pathway, respectively (39 - 42). Meanwhile, IQGAP2 has been identified by mass spectrometry and bioinformatics to possibly participate in the TNF-α-stimulated NF-κB pathway (43). As a transactivator, Rta is often expressed in the nucleus to affect gene transcription. Here, we showed that Rta-induced IQGAP2 associates with Rta and is colocalized with Rta in the nucleus. Nuclear expression of IQGAP2 had not been previously reported. Here, we demonstrated for the first time that Rta may be able to recruit IQGAP2 to the nucleus, where IQGAP2 possibly participates in Rta-related gene regulation.
Lastly, from a medical perspective, elevated antibodies against EBV lytic proteins and increased EBV DNA copy number in the serum have been reported in patients suffering from Burkitt’s lymphoma, Hodgkin’s lymphoma, PTLD, or NPC (2), indicating that EBV lytic progression may play an important role in EBV pathogenesis. Having identified a positive correlation between IQGAP2 and EBV lytic progression in the presence of Rta using our ex vivo EBV-immortalized LCL model, we wanted to further assess how IQGAP2 affects the clinical outcomes of EBV-associated diseases. In order to understand the correlation between IQGAP2 expression and survival, we (data not shown) and another group (44) analyzed the survival data of patients with DLBCL from GEO data sets (accession number GSE10846). Based on the median expression level of IQGAP2, 414 patients were divided into two clusters, high (n = 211) and low (n = 203) IQGAP2 groups. Clinical statistics showed that patients with high IQGAP2 had worse overall survival (P = 0.0007). According to these results, IQGAP2 is not only expressed in EBV-associated diseases but also correlated with worse disease outcomes.
MATERIALS AND METHODS
B cell purification and EBV infection
Peripheral blood mononuclear cells (PBMCs) were isolated from the whole blood of healthy donors by Ficoll-Hypaque density gradient centrifugation (GE Healthcare, Chicago, IL, USA). CD19+ B cells were purified from PBMCs using Dynabeads (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. CD19+ B cells were seeded at a density of 1 × 106 cells/mL in 12-well plates and infected with an EBV stock (dilution 1:100) at a multiplicity of infection (MOI) of 10. Cells were harvested for RNA and protein collection on days 0, 3, 7, 14, 21, and 28 post-infection.
Cell culture
The EBV-positive Akata cell line and EBV-negative Akata cell line were derived from Burkitt’s lymphoma (27). The BJAB cell line was derived from an EBV-negative Burkitt’s lymphoma (27). The TW01 cell line was derived from an EBV-negative nasopharyngeal carcinoma (27). LCLs were established by EBV infection of CD19+ B cells. Akata cells, BJAB cells, and LCLs were cultured in Roswell Park Memorial Institute medium, while 293T, TW01, and NA cells were cultured in Dulbecco’s Modified Eagle Medium. The cell culture media were supplemented with 10% FBS, 1 mM L-glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin. NA cells were treated with 40 ng of TPA per mL and 3 mM SB for the chemical induction of EBV lytic progression.
Electroporation
BJAB cells were electroporated using the Neon Kit (Invitrogen) according to the manufacturer’s instructions. In brief, cells were washed with EDTA-free PBS three times, resuspended, and mixed with the relevant plasmids before electroporation. After pulse application, the samples were transferred into a fresh medium and then incubated for the specified amount of time before being subjected to further assays. Notably, 0.5 µg of pEGFP-C1 or pEGFP-Rta was added to 2 × 105 cells for transfection by electroporation using a pulse of 1,250 V for 30 ms.
RNA extraction and RT-qPCR
Total cellular RNA was extracted with TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. Reverse transcription (RT) and quantitative polymerase chain reaction (qPCR) were carried out as previously described (14). RT was performed with random hexamers and M-MLV Reverse Transcriptase (Promega, Madison, WI, USA). qPCR was performed using the SensiFASTTM SYBR No-ROX Kit (Bioline, London, UK) according to the user manual. ProZyme Tbr DNA polymerase was used in PCR. The primers used for mRNA detection were as follows: IQGAP1: forward primer 5′-GCCAGACAGCACTGTGTTG-3′, reverse primer 5′-TCACGGATAGCACGTCTCTG-3′; IQGAP2: forward primer 5′-CCTTGTGAAGGCAAAAGAGC-3′, reverse primer 5′-CCGCCTGTGTGCATATACTCCT-3′; IQGAP3: forward primer 5′-ATGACTCCAACACCCGTAGC-3′, reverse primer 5′-ACTAGCCCCTGGTAGCCATT-3′; Rta: forward primer 5′-CGGGATCCAAATAGACAGCCCAGTTGAAA-3′, reverse primer 5′-CGGGATCCCAAGAGAGCGATGAGAGAC-3′; Zta: forward primer 5′-TTCCACAGCCTGCACCAGTG-3′, reverse primer 5′-GGCAGCAGCCACCTCACGGT-3′; EBNA1: forward primer 5′-ATGAGCGTTTGGGAGAGCTGATTC-3′, reverse primer 5′-TCCTCGTCCATGGTTATCAC-3′; β-actin: forward primer 5′ -TTCTACAATGAGCTGCGTGT-3′, reverse primer 5′ -GCCAGACAGCACTGTGTTGG-3′.
Plasmid construction
Insert genes were prepared by PCR with Phusion Hot Start High-Fidelity DNA Polymerase with specialized primers and LCL genomic DNA template. DNA plasmids were prepared with QIAGEN Plasmid Plus Midi Kit or Presto Mini Plasmid kit according to the user manuals. The various serial 5′-deleted pGL3-pIQGAP2 plasmids were constructed by inserting IQGAP2 promoter fragments (−853/+291, –642/+291, –523/+291, –167/+291, and −41/+291) into pGL3 Luciferase Reporter Vectors at XhoI and NheI sites. The various pEGFP-Rta plasmids expressing full-length and truncated forms of Rta (1-441, 1-333, and 401-605) were gifts from Dr. Tsuey-Ying Hsu. The Rta-expressing plasmid pSG5-Rta was constructed as described in a previous study (45).
Western blotting and antibodies
Western blotting was performed as described in a previous study (27). Antibodies against β-actin (clone: AC-15; Sigma-Aldrich, Burlington, MA, USA), IQGAP1 (clone: 05-504, Millipore, Burlington, MA, USA), IQGAP2 (clone: BB9, Millipore; clone: H-209, Santa Cruz Biotechnology, Dallas, TX, USA), PARP-1 (clone: F-2, Santa Cruz Biotechnology), ⍺-tubulin (clone: DM1A, Calbiochem, San Diego, CA, USA), E-cadherin (clone: 34, BD Biosciences, San Jose, CA, USA), and mouse and rabbit IgG (Jackson Immunoresearch, West Grove, PA, USA; Cappel Laboratories, Cochranville, PA, USA) were used. Antibodies against Zta (clone: 1B4), Rta (clone: 467 and clone: 37 1H10), EBNA1 (patient’s serum NPC47), EA-D (clone: 88), and LMP1 (clone: S12) were prepared as reported previously (27). Monoclonal mouse IgG and rabbit IgG purified in our lab were also used.
Lentivirus packaging and infection
The methods for lentivirus packaging and infection were similar to those described in a previous study (27). In brief, p8.91, pMD2.G, and shRNA-expressing plasmids were co-transfected into 293T cells using Lipofectamine 2000 (Invitrogen). pLKO.1-shLuciferase, pLKO.1-shIQGAP2-2, and pLKO.1-shIQGAP2-3 are shRNA-expressing plasmids targeting the coding sequence of luciferase or IQGAP2. After 16 h post-transfection, the medium was replaced, and the transfected cells were incubated for another 48 h. Supernatants containing the infectious shRNA lentiviruses were then collected and stored at −80℃ until use. For lentivirus infection, cells were infected at an MOI of 1. Infected cells then underwent selection with 2 µg/mL puromycin for 5 d post-infection before further analysis.
Indirect immunofluorescence assay
An amount of 1 × 104 TW01 cells were seeded on a sterile 22 × 22 mm glass coverslip one night before transfection. pcDNA3 or pcDNA3-IQGAP2-HA, and pEGFP-C1 or pEGFP-Rta were transfected into TW01 cells. The coverslip was collected 72 h post-transfection. For suspension cells, PBS-diluted cells were air dried to be fixed on a glass slide. TW01 cells were cross-linked with 4% formaldehyde, while human CD19+ uninfected B lymphocytes and EBV-immortalized LCLs were fixed with 2% formaldehyde. 0.1% Triton X-100 was used for cell permeabilization before primary antibody incubation with mouse anti-Rta (clone: 467) and rabbit anti-IQGAP2 (clone: H-209, Santa Cruz Biotechnology, Dallas, TX, USA) antibodies. Afterward, FITC-conjugated goat anti-mouse IgG and rhodamine-conjugated goat anti-rabbit IgG (Cappel Laboratories, Cochranville, PA, USA) antibodies were added to the cells, and the nuclei were stained with Hoechst 33342. Fluorescence was detected by fluorescence microscopy or confocal microscopy.
Analysis of Rta- and IQGAP2-positive LCL cells by flow cytometry
Intracellular staining of Rta and IQGAP2 expression was assayed using the protocol of the Foxp3/transcription factor staining buffer set (eBioscience) according to the manufacturer’s instructions. Fixed cells were stained with irrelevant monoclonal mouse IgG and rabbit IgG purified in our lab, or mouse anti-Rta (clone: 37 1H10) and rabbit anti-IQGAP2 (clone: H-209, Santa Cruz Biotechnology, Dallas, TX, USA) antibodies for 30 min at room temperature. The cells were washed and incubated with FITC-conjugated goat anti-mouse IgG and rhodamine-conjugated goat anti-rabbit IgG (Cappel Laboratories, Cochranville, PA, USA) antibodies for 30 min, and then cells were washed again and resuspended in PBS and analyzed by flow cytometry with the BD LSRFortessa Cell Analyzer (BD Biosciences).
Subcellular fractionation
First, 3 × 105 TW01 cells were transfected using NTR II Transfection Reagent (T-pro Biotechnology, Taipei, Taiwan). TW01 cells were transfected with pEGFP-C1 or pEGFP-Rta, and pcDNA3 or pcDNA3-IQGAP2-HA. And 72 h post-transfection, transfected cells were harvested and lysed. The supernatant was harvested as the cytosolic fraction, and the pellets were washed, treated with nuclei lysis buffer (pH 8.0, 50 mM Tris, 10 mM EDTA, 1% SDS, supplemented with 1 mM DTT, 1× protease inhibitor, 0.1 mM PMSF, 0.1 M NaF, 1 mM Na3VO4), homogenized, and centrifuged to obtain the nuclear fraction.
Co-immunoprecipitation
Two types of cells were used in this assay. 293T cells were transfected with pEGFP-C1 or pEGFP-Rta, and pcDNA3-IQGAP2-HA for 72 h and harvested. For immunoprecipitation, cell lysates pre-cleared with Protein A beads (20% slurry) were incubated with an anti-GFP antibody (clone: M2, Sigma-Aldrich) or anti-HA antibody (clone: 16B12, Babco BioLegend, San Diego, CA, USA). LCLs were cultured in RPMI supplemented with 5% FBS for 5 d. Cell lysates were pre-cleared with irrelevant monoclonal antibody and incubated with an anti-Rta antibody (clone: 37 1H10). Protein A beads were added to precipitate the immunocomplexes. After PBS wash, the precipitated immunocomplexes were analyzed by western blotting.
Luciferase reporter assay
293T cells were seeded at a density of 2 × 105 cells/well in 12-well plates and then transfected with 0.5 µg of the relevant luciferase reporter plasmid, 0.05 µg of the GFP-expressing plasmid pEGFP-C1 (Promega), and 0.5 µg of the relevant effector plasmid using NTR II. After 72 h post-transfection, the cells were lysed, and the luciferase activities and GFP fluorescence intensities were detected using the Bright-Glo Luciferase Assay System (Promega, Madison, WI, USA), Orion II Microplate Luminometer (Titertek-Berthold, Pforzheim, Germany), and SpectraMax M5 Microplate Reader (Molecular Devices, San Jose, CA, USA). The luciferase activity of each transfectant was normalized to its GFP fluorescence intensity and standardized using the normalized luciferase activity of the vector control cells to obtain the fold change.
Chromatin immunoprecipitation assay
5 × 106 293 T cells were transfected with pEGFP-C1 or pEGFP-Rta using NTR II as previously mentioned in the section describing subcellular fractionation. And 72 h post-transfection, cells were cross-linked with 1% formaldehyde. Afterward, cells were lysed with nuclei lysis buffer and sonicated to generate DNA fragments between 500 and 1,000 bps in length. After centrifugation, the supernatants were kept and their DNA-protein complex concentrations were determined. An amount of 200 µg of the DNA-protein complexes from each supernatant was then obtained for immunoprecipitation with anti-GFP or control mouse IgG antibody. The immunocomplexes were captured by Protein A beads, and the beads were then washed twice with 1 mL of low salt buffer (pH 8.0, 50 mM Tris, 150 mM NaCl, 5 mM EDTA, 1% IGEPAL, 0.1% SDS, 1% deoxycholic acid), 1 mL of high salt buffer (pH 8.0, 50 mM Tris, 300 mM NaCl, 5 mM EDTA, 1% IGEPAL, 0.1% SDS, 1% deoxycholic acid), and 1 mL of LiCl buffer (pH 8.0, 50 mM Tris, 150 mM NaCl, 5 mM EDTA, 300 mM LiCl, 1% IGEPAL, 0.1% SDS, 1% deoxycholic acid) in the order listed. To de-cross-link the DNA-protein complexes, elution buffer (50 mM NaHCO3 and 1% SDS) was added to the beads, and the reaction mixtures were incubated at 65°C overnight. The reaction mixtures were centrifuged, and the supernatants were collected and treated with 10 mg/mL of proteinase K. The DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and then precipitated with absolute ethanol and 20 µg of glycogen. The DNA pellet was washed with 70% ethanol and dissolved in H2O. Two microliters of DNA from each sample were taken as a template for PCR to detect the fragment containing the IQGAP2 promoter region −232/+65. The three temperature steps were set at 95°C for 1 min, 55°C for 30 s, and 72°C for 30 s, and repeated for a total of 40 cycles. Finally, the PCR products were detected in an EtBr-stained 2% agarose gel under UV irradiation.
Cell proliferation rate determination
Lentivirus infection of LCLs was performed as previously mentioned in the section describing lentivirus packaging and infection. Cells were seeded at a density of 1 × 104 cells/well in 96-well plates and treated with alamarBlue (AbD Serotec, Kidlington, UK). After incubation for 4 h, absorbance at 570 nm and 600 nm was measured to obtain the percentage reduction of alamarBlue according to the user manual.
ACKNOWLEDGMENTS
This work was supported by the National Science and Technology Council (109-2320-B-002-052-MY3).
K.M.L. designed and performed the experiments, analyzed the data, and co-wrote the manuscript; L.F.W. designed and performed the experiments, analyzed the data, and co-wrote the manuscript; S.Y.C. designed and performed the experiments, analyzed the data, and co-wrote the manuscript; S.J.L. designed and performed the experiments, analyzed the data, and co-wrote the manuscript; and C.H.T. designed the experiments and co-wrote the manuscript.
All authors declare that they have no competing financial interests.
Contributor Information
Sue-Jane Lin, Email: suejane.lin@gmail.com.
Ching-Hwa Tsai, Email: chtsai@ntu.edu.tw.
Lori Frappier, University of Toronto, Toronto, Ontario, Canada .
REFERENCES
- 1. Macsween KF, Crawford DH. 2003. Epstein-Barr virus-recent advances. Lancet Infect Dis 3:131–140. doi: 10.1016/s1473-3099(03)00543-7 [DOI] [PubMed] [Google Scholar]
- 2. Young LS, Yap LF, Murray PG. 2016. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 16:789–802. doi: 10.1038/nrc.2016.92 [DOI] [PubMed] [Google Scholar]
- 3. Sausen DG, Bhutta MS, Gallo ES, Dahari H, Borenstein R. 2021. Stress-induced Epstein-Barr virus reactivation. Biomolecules 11:1380. doi: 10.3390/biom11091380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kerr JR. 2019. Epstein-Barr virus (EBV) reactivation and therapeutic inhibitors. J Clin Pathol 72:651–658. doi: 10.1136/jclinpath-2019-205822 [DOI] [PubMed] [Google Scholar]
- 5. Martinez OM, Krams SM. 2017. The immune response to Epstein Barr virus and implications for posttransplant lymphoproliferative disorder. Transplantation 101:2009–2016. doi: 10.1097/TP.0000000000001767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simonnet A, Engelmann I, Moreau A-S, Garcia B, Six S, El Kalioubie A, Robriquet L, Hober D, Jourdain M. 2021. High incidence of Epstein-Barr virus, cytomegalovirus, and human-herpes Virus-6 reactivations in critically ill patients with COVID-19. Infect Dis Now 51:296–299. doi: 10.1016/j.idnow.2021.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amon W, Farrell PJ. 2005. Reactivation of Epstein-Barr virus from latency. Rev Med Virol 15:149–156. doi: 10.1002/rmv.456 [DOI] [PubMed] [Google Scholar]
- 8. Gruffat H, Marchione R, Manet E. 2016. Herpesvirus late gene expression: a viral-specific pre-initiation complex is key. Front Microbiol 7:869. doi: 10.3389/fmicb.2016.00869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adamson AL, Darr D, Holley-Guthrie E, Johnson RA, Mauser A, Swenson J, Kenney S. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated P38 and C-Jun N-terminal kinases. J Virol 74:1224–1233. doi: 10.1128/jvi.74.3.1224-1233.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu P, Speck SH. 2003. Synergistic autoactivation of the Epstein-Barr virus immediate-early BRLF1 promoter by RTA and Zta. Virology 310:199–206. doi: 10.1016/s0042-6822(03)00145-4 [DOI] [PubMed] [Google Scholar]
- 11. Kenney SC, Mertz JE. 2014. Regulation of the latent-lytic switch in Epstein-Barr virus. Semin Cancer Biol 26:60–68. doi: 10.1016/j.semcancer.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heilmann AMF, Calderwood MA, Portal D, Lu Y, Johannsen E. 2012. Genome-wide analysis of Epstein-Barr virus Rta DNA binding. J Virol 86:5151–5164. doi: 10.1128/JVI.06760-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ho CH, Hsu CF, Fong PF, Tai SK, Hsieh SL, Chen CJ. 2007. Epstein-Barr virus transcription activator Rta Upregulates decoy receptor 3 expression by binding to its promoter. J Virol 81:4837–4847. doi: 10.1128/JVI.02448-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chua HH, Lee HH, Chang SS, Lu CC, Yeh TH, Hsu TY, Cheng TH, Cheng JT, Chen MR, Tsai CH. 2007. Role of the TSG101 gene in Epstein-Barr virus late gene transcription. J Virol 81:2459–2471. doi: 10.1128/JVI.02289-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang LK, Chung JY, Hong YR, Ichimura T, Nakao M, Liu ST. 2005. Activation of Sp1-mediated transcription by Rta of Epstein-Barr virus via an interaction with MCAF1. Nucleic Acids Res 33:6528–6539. doi: 10.1093/nar/gki956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ragoczy T, Miller G. 2001. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J Virol 75:5240–5251. doi: 10.1128/JVI.75.11.5240-5251.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hedman AC, Smith JM, Sacks DB. 2015. The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep 16:427–446. doi: 10.15252/embr.201439834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Briggs MW, Sacks DB. 2003. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep 4:571–574. doi: 10.1038/sj.embor.embor867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dai Q, Ain Q, Rooney M, Song F, Zipprich A. 2022. Role of IQ motif-containing GTPase-activating proteins in hepatocellular carcinoma. Front Oncol 12:920652. doi: 10.3389/fonc.2022.920652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xie Y, Zheng L, Tao L. 2019. Downregulation of IQGAP2 correlates with prostate cancer recurrence and metastasis. Transl Oncol 12:236–244. doi: 10.1016/j.tranon.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar D, Patel SA, Khan R, Chawla S, Mohapatra N, Dixit M. 2022. IQ motif-containing GTPase-activating protein 2 inhibits breast cancer angiogenesis by suppressing VEGFR-AKT signaling. Mol Cancer Res 20:77–91. doi: 10.1158/1541-7786.MCR-20-1044 [DOI] [PubMed] [Google Scholar]
- 22. Xu L, Shao Y, Ren L, Liu X, Li Y, Xu J, Ye Y. 2020. IQGAP2 inhibits migration and invasion of gastric cancer cells via elevating SHIP2 phosphatase activity. Int J Mol Sci 21:1968. doi: 10.3390/ijms21061968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu J, Qu Y, Liu Y, Jambusaria R, Han Z, Ruthel G, Freedman BD, Harty RN. 2013. Host IQGAP1 and Ebola virus VP40 interactions facilitate virus-like particle egress. J Virol 87:7777–7780. doi: 10.1128/JVI.00470-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gladue DP, Holinka LG, Fernandez-Sainz IJ, Prarat MV, O’Donnell V, Vepkhvadze NG, Lu Z, Risatti GR, Borca MV. 2011. Interaction between core protein of classical swine fever virus with cellular IQGAP1 protein appears essential for virulence in swine. Virology 412:68–74. doi: 10.1016/j.virol.2010.12.060 [DOI] [PubMed] [Google Scholar]
- 25. Leung J, Yueh A, Appah FSK, Yuan B, de los Santos K, Goff SP. 2006. Interaction of moloney murine leukemia virus matrix protein with IQGAP. EMBO J 25:2155–2166. doi: 10.1038/sj.emboj.7601097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brisac C, Salloum S, Yang V, Schaefer EAK, Holmes JA, Chevaliez S, Hong J, Carlton-Smith C, Alatrakchi N, Kruger A, Lin W, Chung RT. 2016. IQGAP2 is a novel interferon-alpha antiviral effector gene acting non-conventionally through the NF-kappaB pathway. J Hepatol 65:972–979. doi: 10.1016/j.jhep.2016.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsai S-C, Lin S-J, Chen P-W, Luo W-Y, Yeh T-H, Wang H-W, Chen C-J, Tsai C-H. 2009. EBV Zta protein induces the expression of Interleukin-13, promoting the proliferation of EBV-infected B cells and Lymphoblastoid cell lines. Blood 114:109–118. doi: 10.1182/blood-2008-12-193375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lai KY, Chou YC, Lin JH, Liu Y, Lin KM, Doong SL, Chen MR, Yeh TH, Lin SJ, Tsai CH. 2015. Maintenance of Epstein-Barr virus latent status by a novel mechanism, latent membrane protein 1-induced Interleukin-32, via the protein kinase Cdelta pathway. J Virol 89:5968–5980. doi: 10.1128/JVI.00168-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gruffat H, Sergeant A. 1994. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res 22:1172–1178. doi: 10.1093/nar/22.7.1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu C, Sista ND, Pagano JS. 1996. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J Virol 70:2545–2555. doi: 10.1128/JVI.70.4.2545-2555.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinson AR, Kwek SS, Hagemeier SR, Wille CK, Kenney SC. 2011. Cellular transcription factor Oct-1 interacts with the Epstein-Barr virus BRLF1 protein to promote disruption of viral latency. J Virol 85:8940–8953. doi: 10.1128/JVI.00569-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gutsch DE, Marcu KB, Kenney SC. 1994. The Epstein-Barr virus BRLF1 gene product transactivates the murine and human c-myc promoters. Cell Mol Biol (Noisy-le-grand) 40:747–760. [PubMed] [Google Scholar]
- 33. Zacny VL, Wilson J, Pagano JS. 1998. The Epstein-Barr virus immediate-early gene product, BRLF1, interacts with the retinoblastoma protein during the viral lytic cycle. J Virol 72:8043–8051. doi: 10.1128/JVI.72.10.8043-8051.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Almasan A, Yin Y, Kelly RE, Lee EY, Bradley A, Li W, Bertino JR, Wahl GM. 1995. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis. Proc Natl Acad Sci U S A 92:5436–5440. doi: 10.1073/pnas.92.12.5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Y, Webster-Cyriaque J, Tomlinson CC, Yohe M, Kenney S. 2004. Fatty acid synthase expression is induced by the Epstein-Barr virus immediate-early protein BRLF1 and is required for Lytic viral gene expression. J Virol 78:4197–4206. doi: 10.1128/jvi.78.8.4197-4206.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xie Y, Yan J, Cutz JC, Rybak AP, He L, Wei F, Kapoor A, Schmidt VA, Tao L, Tang D. 2012. IQGAP2, A candidate tumour suppressor of prostate tumorigenesis. Biochim Biophys Acta 1822:875–884. doi: 10.1016/j.bbadis.2012.02.019 [DOI] [PubMed] [Google Scholar]
- 37. Jin SH, Akiyama Y, Fukamachi H, Yanagihara K, Akashi T, Yuasa Y. 2008. IQGAP2 inactivation through aberrant promoter methylation and promotion of invasion in gastric cancer cells. Int J Cancer 122:1040–1046. doi: 10.1002/ijc.23181 [DOI] [PubMed] [Google Scholar]
- 38. Schmidt VA, Scudder L, Devoe CE, Bernards A, Cupit LD, Bahou WF. 2003. IQGAP2 functions as a GTP-dependent effector protein in thrombin-induced platelet cytoskeletal reorganization. Blood 101:3021–3028. doi: 10.1182/blood-2002-09-2807 [DOI] [PubMed] [Google Scholar]
- 39. Cheung KL, Lee JH, Shu L, Kim J-H, Sacks DB, Kong A-NT. 2013. The Ras GTPase-activating-like protein IQGAP1 mediates Nrf2 protein activation via the mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)-ERK pathway. J Biol Chem 288:22378–22386. doi: 10.1074/jbc.M112.444182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim J-H, Xu EY, Sacks DB, Lee J, Shu L, Xia B, Kong A-N. 2013. Identification and functional studies of a new Nrf2 partner IQGAP1: a critical role in the stability and transactivation of Nrf2. Antioxid Redox Signal 19:89–101. doi: 10.1089/ars.2012.4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, Schmidt V, Hogan PG, Sacks DB, Rao A. 2011. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A 108:11381–11386. doi: 10.1073/pnas.1019711108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Erdemir HH, Li Z, Sacks DB. 2014. IQGAP1 binds to estrogen receptor-alpha and modulates its function. J Biol Chem 289:9100–9112. doi: 10.1074/jbc.M114.553511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bouwmeester T, Bauch A, Ruffner H, Angrand P-O, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon A-M, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin A-C, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. 2004. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6:97–105. doi: 10.1038/ncb1086 [DOI] [PubMed] [Google Scholar]
- 44. Tang T, Wang J, Zhang L, Cheng Y, Saleh L, Gu Y, Zhang H. 2021. IQGAP2 acts as an independent prognostic factor and is related to immunosuppression in DLBCL. BMC Cancer 21:603. doi: 10.1186/s12885-021-08086-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chang Y, Lee HH, Chang SS, Hsu TY, Wang PW, Chang YS, Takada K, Tsai CH. 2004. Induction of Epstein-Barr virus latent membrane protein 1 by a lytic transactivator Rta. J Virol 78:13028–13036. doi: 10.1128/JVI.78.23.13028-13036.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]