Fig 1.
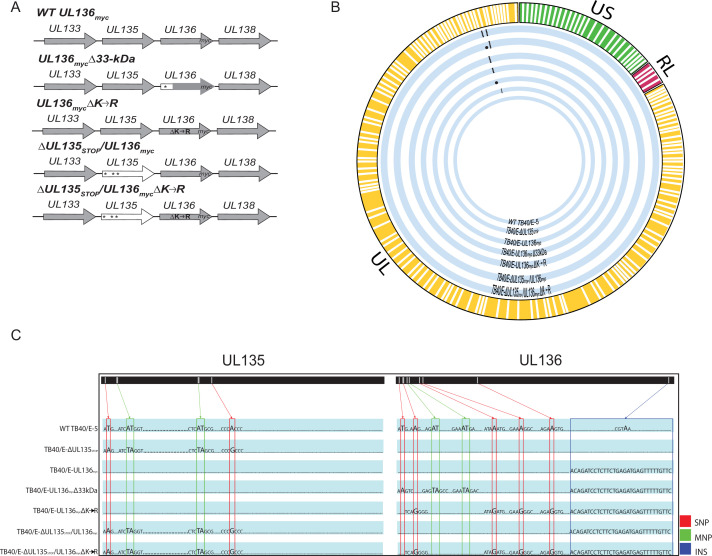
Schematic and whole-genome sequencing of viruses used in this study. (A) Schematic of changes within the UL133–UL138 locus in TB40/E recombinant viruses indicated. Parental (WT) UL136myc expresses UL136 fused in frame with a C-terminal myc epitope tag. All recombinant viruses used in this study are derived from this virus and express the myc epitope-tagged version of UL136 for protein detection. UL136mycΔ33kDa contains stop codon substitutions for 5’ AUGs to disrupt the expression of UL136p33. UL136mycΔK→R contains arginine substitutions for all four lysine residues in UL136 (amino acid positions 4, 20, 25, and 113). ΔUL135STOP/ΔUL136myc contains stop codon substitutions for 5’ AUGs to abrogate synthesis of UL135 protein (amino acid positions 1, 27, and 97). ΔUL135STOP/ΔUL136mycΔK→R combines the stop codon substitutions in UL135 with the lysine to arginine substitutions in UL136 described above. * indicates stop codon substitutions for methionine codons. (B) Whole-genome sequencing and alignment of parental (TB40/E) and recombinant HCMV bacterial artificial chromosomes (BACs) was performed to identify alterations in each recombinant virus. Thick blue lines represent each recombinant virus as labeled, and mutations in each genome are denoted by black dots. (C) Zoomed in pictograph of UL135 and UL136 genes with specific nucleotide changes evident in sequencing. Single-nucleotide polymorphisms (SNPs) are boxed in red; multiple nucleotide polymorphisms (MNPs) are boxed in green; and insertions (INS) are boxed in blue. Mutations for each recombinant virus are compared back to the WT TB40/E-5 parental virus.