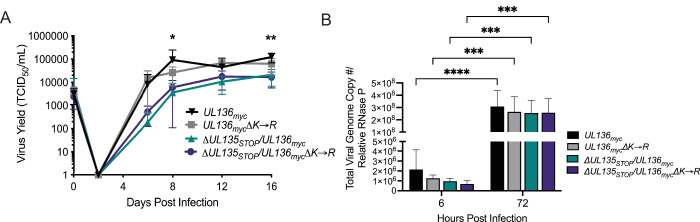
Fig 3.
Stabilization of UL136p33 does not rescue viral yields associated with disruption of UL135 in productive infection. (A) Primary MRC-5 cells were infected with WT UL136 myc, UL136mycΔK→R, ΔUL135STOP/UL136myc, or ΔUL135STOP/UL136mycΔK→R recombinant viruses, and a multistep (MOI of 0.02) growth curve was performed. Cells and culture supernatant were collected at the indicated time points, and virus titers were measured by TCID50. Data points represent the averages from three independent experiments and error bars represent standard deviations. A two-way ANOVA with Tukey’s multiple comparison tests were performed to determine statistical significance for each mutant infection relative to the WT UL136myc infection. *P < 0.05 for WT UL136myc compared to ΔUL135STOP/UL136myc and ΔUL135 STOP/UL136mycΔK→R at 8 dpi and **P < 0.01 for WT UL136myc compared to ΔUL135STOP/UL136myc and ΔUL135STOP/UL136mycΔK→R at16 dpi. (B) Total DNA was isolated from primary MRC-5 cells infected at an MOI of 1 with WT UL136 myc, UL136mycΔK→R, ΔUL135STOP/UL136myc, or ΔUL135STOP/UL136mycΔK→R recombinant viruses at 6 and 72 hpi. The total number of viral genomes were quantified using qPCR with primers to β2.7kb relative to the cellular RNaseP gene. The viral genomes present in the cell at 6 hpi represent the input genome copy number. Averages of duplicate measurements from three independent experiments with standard deviation bars are shown for each virus at each time point. A two-way ANOVA with Tukey’s multiple comparison tests were performed to determine statistical significance values for each mutant infection relative to its 6 hpi time point (***, P < 0.001 and ****, P < 0.0001).