Abstract
Germline antibodies, the initial set of antibodies produced by the immune system, are critical for host defense, and information about their binding properties can be useful for designing vaccines, understanding the origins of autoantibodies, and developing monoclonal antibodies. Numerous studies have found that germline antibodies are polyreactive with malleable, flexible binding pockets. While insightful, it remains unclear how broadly this model applies, as there are many families of antibodies that have not yet been studied. In addition, the methods used to obtain germline antibodies typically rely on assumptions and do not work well for many antibodies. Herein, we present a distinct approach for isolating germline antibodies that involves immunizing activation-induced cytidine deaminase (AID) knockout mice. This strategy amplifies antigen-specific B cells, but somatic hypermutation does not occur because AID is absent. Using synthetic haptens, glycoproteins, and whole cells, we obtained germline antibodies to an assortment of clinically important tumor-associated carbohydrate antigens, including Lewis Y, the Tn antigen, sialyl Lewis C, and Lewis X (CD15/SSEA-1). Through glycan microarray profiling and cell binding, we demonstrate that all but one of these germline antibodies had high selectivity for their glycan targets. Using molecular dynamics simulations, we provide insights into the structural basis of glycan recognition. The results have important implications for designing carbohydrate-based vaccines, developing anti-glycan monoclonal antibodies, and understanding antibody evolution within the immune system.
Graphical Abstract

INTRODUCTION
Antibodies are a critical component of the mammalian immune system and are valuable tools for basic research and clinical applications. Their key function is to bind pathogens, foreign materials, or other target molecules and either directly neutralize them or tag them for destruction by the immune system.1 The immune system produces a large and diverse repertoire of antibodies that changes over time. The initial antibodies produced by naïve B cells are unmutated and referred to as “germline antibodies”. They are the first to be produced in response to antigen exposure or infection and play an important role in the primary immune response. Subsequently, mutations are introduced into germline antibodies via somatic hypermutation, and mutants with improved affinity are enriched through a repetitive selection process known as affinity maturation. “Affinity matured” antibodies provide more potent protection and are typically the antibodies pursued for clinical applications.2 Thus, in addition to contributing to immune defense, germline antibodies also serve as precursors for affinity-matured antibodies.
Given the importance of germline antibodies, an in-depth understanding of their binding properties is useful for a variety of reasons. It provides fundamental knowledge regarding the capacity of the initial antibody repertoire to recognize diverse antigens and protect us from a plethora of continually evolving pathogens. Knowledge of germline binding properties is also valuable for designing vaccines and immunogens that are capable of inducing a strong and productive antibody response.3 Furthermore, comparison of affinity-matured antibodies with their corresponding germline precursors provides insights as to how various mutations influence affinity and selectivity and can facilitate engineering monoclonal antibodies with improved properties.
Numerous prior studies have evaluated the binding properties, structures, and dynamics of germline antibodies.4–23 In general, this body of work has demonstrated that germline antibodies are polyreactive/polyspecific relative to their affinity-matured counterparts. A proposed model explaining this observation, referred to as the polyspecificity hypothesis, suggests that the germline encodes structurally flexible antibodies capable of recognizing a range of antigenic structures with low to modest affinity. During affinity maturation, mutations preorganize or rigidify the binding pocket and optimize contacts between the antibody and antigen. Polyspecificity is an appealing concept as it significantly expands the number of antigens recognized by the immune system; however, very few germline antibodies have been studied in detail, and so, it is unclear the extent to which this model applies. Germline antibodies with high selectivity are plausible, but evidence for these antibodies is limited.24
A major challenge in the field is identifying germline antibodies, especially to specific antigens of interest. Most commonly, the germline antibody precursor is inferred from the sequence of the affinity-matured antibody using database programs such as IgBLAST.25,26 This process works by aligning the sequence of interest with the known V, D, and J germline genes used to construct antibodies (Figure 1) and identifying the most similar matches. While useful, this strategy relies on various assumptions and does not work well in many cases.27 As examples, the D gene is often too short to reliably determine, and B cells can insert random, nontemplated nucleotides in the V–D and D–J junctions, which cannot be assigned via alignments.27 As a result, the true germline sequence in this region can be very difficult to determine. A second approach for obtaining germline antibodies involves cloning antibody genes from naïve B cells.28–30 While this approach has certain advantages, it can be difficult to obtain naïve B cells that bind to a specific target of interest or identify the natural target(s) for any given naïve B cell. Consequently, it is nontrivial to obtain matched antigen-germline antibody pairs with this approach. Furthermore, methods used to isolate naïve B cells do not achieve 100% purity; thus, some antibodies evaluated in this process may be derived from other B cells and mistaken for germline antibodies.
Figure 1.

Assembly of antibody genes. Antibodies are composed of two light chains and two heavy chains. The sequences for these chains are assembled via recombination of V, D, and J genes for the heavy chain and V and J genes for the light chain. The D genes are relatively short and can be trimmed, often resulting in lengths of only 5–15 nucleotides. Diversity is further increased by insertion of nontemplated nucleotides in the V–D and D–J junctions of the heavy chain.
In this study, we describe a unique strategy to obtain germline monoclonal antibodies from mice lacking the key enzyme required for somatic hypermutation, activation-induced cytidine deaminase (AID or AICDA). AID knockout mice are capable of mounting an antibody response, but they do so without somatic hypermutation or isotype switching.31 Prior to this work, it was not known if the responses in AID KO mice would be robust enough to generate monoclonal antibodies, especially for antigens with low immunogenicity such as tumor-associated carbohydrate antigens. Herein, we demonstrate the successful use of this approach in isolating germline antibodies to a variety of clinically relevant carbohydrate antigens. Rather than being polyreactive, nearly all of the glycan-binding germline antibodies were highly selective for their target antigen, supporting a distinct model for the development of anti-glycan antibodies within the mammalian immune system.
RESULTS
Isolation of Germline-Encoded Monoclonal Antibodies to Lewis Y.
As a first test, our goal was to generate germline antibodies to Lewis Y, a tumor-associated carbohydrate antigen that is highly overexpressed on a variety of tumors. Antibodies to Lewis Y have progressed into clinical trials as single agents, antibody–drug conjugates, radio-immunotherapies and as components of CAR T cells.32–42 To induce responses to Lewis Y, AID knockout C57BL/6 mice31 were immunized with a Lewis Y hapten-human serum albumin conjugate (LeY-tetra-HSA; Fucα1–2Galβ1–4(Fucα1–3)GlcNAc-HSA). The conjugate was prepared by reductive amination of a Lewis Y terminal pentasaccharide [Fucα1–2Galβ1–4(Fucα1–3)GlcNAcβ1–3Gal] with human serum albumin (Figure 2A). In this process, the reducing end galactose undergoes ring opening and attachment to an amine side chain of albumin; therefore, we consider it part of the linker, rather than part of the glycan structure.
Figure 2.

Structures of selected Lewis Y and related conjugates. (A) Lewis Y hapten conjugates were prepared via reductive amination to free amines on albumin (lysine side chains and/or the N-terminus). (B) Glycans and linkers for selected array components.
Standard procedures were then used to fuse spleen B cells with myeloma cells to generate hybridomas. These hybridomas were distributed in 96-well plates as oligoclonal mixtures (estimated 1–6 clones per well), and the resulting supernatants were screened by ELISA for binding to Lewis Y-tetra-10-BSA. With the goal of obtaining a range of antibodies and minimizing bias, we selected 25 wells that gave signals (both weak and strong) to profile on our glycan microarray.43–47 Our microarray was composed of a diverse collection of N-linked and O-linked glycans, glycans from glycolipids, glycopeptides, nonhuman glycans, and glycoproteins.48–50 It contained numerous glycans relevant for this study. First, it had three different glycan components with a terminal Lewis Y tetrasaccharide (Figure 2B). “LeY-08” and “LeY-tetra” contain the same tetrasaccharide but have different linkers. “LeY-LeX” is a heptasaccharide with a terminal Lewis Y appended to a Lewis X carrier glycan chain. In addition to the glycans with a terminal Lewis Y, our array also had two glycans that have an internal Lewis Y structure: 2′F-A type 2-Sp [GalNAcα1–3(Fucα1–2)Galβ1–4(Fucα1–3)GlcNAc] and 2′F-B type 2-Sp [Galα1–3(Fucα1–2)Galβ1–4(Fucα1–3)GlcNAc]. Several glycans that are substructures of Lewis Y were also included: Lewis X [LeX; Galβ1–4(Fucα1–3)GlcNAc], blood group H2 [BG-H2; Fucα1–2Galβ1–4GlcNAc], blood group H (BG-H; Fucα1–2Gal), and LacNAc (Galβ1–4GlcNAc). In addition, the array contained many other Lewis antigens, including Lewis A, Lewis B, Lewis C, Sialyl Lewis A, and Sialyl Lewis X. Lastly, the array contained double stranded DNA (dsDNA) and dinitrophenyl-modified BSA (DNP-BSA), two molecules often bound by polyreactive antibodies.28 Most of the oligoclonal supernatants that were profiled displayed selective binding to Lewis Y or binding to Lewis Y and fucose; however, a few had signals to other array components, including DNP-BSA. Because there can be several different antibodies contributing to signals, we could not determine if these were polyreactive antibodies at this stage.
We selected a variety of wells for subcloning and successfully obtained seven Lewis Y reactive clones. Four came from wells where signals were only observed to Lewis Y. After sequencing, all four were found to be identical and designated as monoclonal antibody 8F11. Two came from wells where signals were observed to Lewis Y and fucose monosaccharide. After sequencing, both were found to be identical and designated 12D6. The last one came from a well that could potentially contain a polyreactive antibody. This monoclonal antibody was designated 8G6. We attempted to subclone another well that could potentially contain a polyreactive antibody and obtained an antibody that bound DNP-BSA, but it did not bind Lewis Y. No further work was carried out on this clone. All monoclonal antibodies were IgM isotype, as AID KO mice do not undergo isotype switching and do not produce IgG antibodies.
Overall, the sequences of the three anti-Lewis Y antibodies were very similar, particularly for the light chains (see Supporting Information and Figure S1). A summary of the germline gene usage can be found in Table 1. For the heavy chain D genes, the IgBLAST database provided multiple potential matches for each antibody, highlighting the difficulties of using sequence similarity to assign germline genes. While we cannot confidently assign the D gene, the AID KO approach ensures that somatic hypermutation has not occurred.
Table 1.
Germline Gene Usage as Determined by IgBLAST25
| heavy chain |
light chain |
|||||
|---|---|---|---|---|---|---|
| glycan | antibody | V | D | J | V | J |
| LeY | 8F11 | IGHV5-12*02 | IGHD1-2*01 (5/5) IGHD2-12*01 (5/5) IGHD2-14*01 (8/9) |
IGHJ3*01 | IGKV1-117*01 | IGKJ4*01 |
| 12D6 | IGHV5-6-3*01 | IGHD2-4*01 (12/12) IGHD2-9*02 (12/12) |
IGHJ3*01 | IGKV1-117*01 | IGKJ2*01 | |
| 8G6 | IGHV5-6-3*01 | IGHD2-4*01 (10/10) IGHD2-9*02 (10/10) |
IGHJ3*01 | IGKV1-117*01 | IGKJ2*01 | |
| H18A, 3S193, BR55, BR96, B3, and B5 | IGHV5-12*02 | IGHJ3*01 | IGKV1-117*01 | IGKJ4*01 | ||
| Tn | 8C11 | IGHV5-17*02 | IGHD1-2*01 (10/10) | IGHJ2*01 | IGKV4-80*01 | IGKJ1*01 |
| 9A9 | IGHV1S53*02 | IGHD3-2*02 (5/5) | IGHJ2*01 | IGKV6-23*01 | IGKJ4*01 | |
| SLeC | 2D3.C11 | IGHV4-1*02 | IGHD4-1*01 (9/9) 1GHD4-1*02 (9/9) |
IGHJ2*01 | IGKV1-110*01 | IGHJ1*01 |
| LeX | 4E9 | M1 BALBc 43 (see Corcoran84) | IGHD1-1*01 (8/8) | IGHJ2*01 | IGKV2-109*01 | IGKJ1*01 |
| 5B3 | IGHV4-1*02 | IGHD1-2*01 (11/11) | IGHJ3*01 | IGKV2-109*01 | IGKJ1*01 | |
| 6D5 | IGHV4-1*02 | IGHD4-1*01 (7/7) IGHD4-1*02 (7/7) |
IGHJ2*01 | IGKV19-93*01 | IGKJ1*01 | |
| 11A11 | 1GHV2-9*02 | IGHD1-2*01 (10/10) | 1GHJ3*01 | IGKV2-109*01 | IGKJ1*01 | |
| 16B9 | 1GHV3-6*02 | Too short to assign | IGHJ2*01 | IGKV6-32*01 | IGKJ4*01 | |
In addition to comparing these sequences to each other, we also compared them to known anti-glycan antibodies. Antibody 8F11 was very similar to six known anti-Lewis Y affinity-matured mouse monoclonal antibodies that have been used in various preclinical and clinical studies for cancer applications (H18A,51 3S193,52 BR55,53 BR96,54 B3,55 and B5;55 Figure 3). All six affinity-matured antibodies share the same inferred V and J genes for both the heavy and light chains, which are identical to the genes used by our germline 8F11 antibody. In addition, all have the same length CDRH3 and >75% sequence identity in the junction region. Given the high degree of sequence similarity, 8F11 may be the true germline precursor for one or more of the six affinity-matured antibodies (see Supporting Information for a more detailed analysis); however, we cannot retroactively confirm whether 8F11 was present in the mice that originally gave rise to these antibodies. The similarity between these antibodies also demonstrates that, at least in some cases, AID KO mice use the same V and J genes as wild-type mice to form anti-Lewis Y antibodies.
Figure 3.

Comparison of anti-Lewis Y antibody sequences. (A) Amino acid sequence alignment of anti-Lewis Y antibodies using 8F11 as the reference sequence. (B) Nucleotide and amino acid sequence of the VDJ junction region of 8F11 compared to H18A/H18A inferred germline (H18A-iGL). The sequence of H18A is identical to the inferred germline for the nucleotides shown. Junctional nucleotides for H18A-iGL are shown in red. Germline V and J gene assignments for 8F11/H18A are provided. IMGT complementary determining regions are shown in color.
Germline Antibodies to Lewis Y Display High Selectivity on a Glycan Microarray.
Each of the antibodies was evaluated on the microarray as an IgM, the first isotype to be produced in an immune response and hence a physiologically relevant isotype for germline antibodies. 8F11, 12D6, and 8G6 were profiled at a range of concentrations on the microarray, and apparent KD values were determined (Table 2 and Figure S2). All three antibodies exhibited apparent KD values of approximately 30 pM for LeY-tetra, a component that had the same glycan and linker as the hapten used for the immunizations (Figure 2A). Because these are IgM antibodies with 10 binding sites, multivalent binding provides very high avidity interactions with our multivalent array surface. In terms of selectivity, 8F11, 12D6, and 8G6 displayed remarkably high selectivity for Lewis Y over other glycans on our microarray (Figures 4A and S3A). Antibody 8F11 was the most selective. Even at the highest antibody concentration tested, approximately 50-fold higher than the apparent KD value for LeY-tetra, binding was only observed to Lewis Y terminal glycans. Antibody 12D6 had the second-best selectivity. It had some recognition of fucose monosaccharide alone (connected to the array via a long, flexible synthetic linker), but it did not bind any other fucose-containing oligosaccharides. Antibody 8G6 also bound fucose monosaccharide and additionally bound blood group H2 (about 15-fold weaker) and blood group H6 (~100-fold weaker). Although the original well that gave rise to 8G6 also displayed binding to Rha-b and DNP-BSA, these signals were eliminated after subcloning. Thus, the signals for Rha-b and DNP-BSA in the oligoclonal well were due to one or more other antibodies present in the mixture.
Table 2.
KD,App of AID KO Germline antibodies (nM)a
| antibody | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| glycan | 8F11 | 12D6 | 8G6 | 8C11 | 9A9 | 2D3.C11 | 4E9 | 5B3 | 6D5 | 11A11 | 16B9 |
| LeY-tetra-10 | 0.039 | 0.024 | 0.036 | – | – | – | – | – | – | – | – |
| LeY-08 | ND | – | – | – | – | – | – | – | – | – | |
| LeY-LeX | ND | – | – | – | – | – | – | – | – | – | – |
| Fuc-α-22 | – | ND | ND | – | – | – | – | – | – | – | – |
| Fuc-β-22 | – | ND | ND | – | – | – | – | – | – | – | – |
| BG-H2-12 | – | – | ND | – | – | – | – | – | – | – | – |
| OSM (Asialo) | – | – | – | 0.042 | 0.039 | – | – | – | – | – | – |
| Ac-Tn(Ser)-S-G-22 | – | – | – | 0.023 | – | – | – | – | – | – | – |
| Ac-S-Tn(Thr)-A-G-22 | – | – | – | 0.018 | – | – | – | – | – | – | – |
| Ac-S-Tn(Thr)-Tn(Thr)-G-09 | – | – | – | 0.45 | 0.032 | – | – | – | – | – | – |
| Tn(Ser)Tn(Ser)Tn(Ser)-G-11 | – | – | – | 0.071 | 0.026 | – | – | – | – | – | – |
| Ac-Tn(Thr)-Tn(Thr)-Tn(Thr)-G-20 | – | – | – | 0.12 | 0.021 | – | – | – | – | – | – |
| GalNac-α-22 | – | – | – | 0.014 | – | – | – | – | – | – | – |
| BG-A-19 | – | – | – | 0.048 | – | – | – | – | – | – | – |
| ForssmanTetra-BSA-13 | – | – | – | 0.027 | – | – | – | – | – | – | – |
| GalNAca1-3Gal | – | – | – | 0.008 | – | – | – | – | – | – | – |
| 3’Neu5Ac-LeC-Sp-12 | – | – | – | – | – | ND | – | – | – | – | – |
| LeX (dimeric)- 07 | – | – | – | – | – | – | ND | 0.008 | 0.039 | 0.050 | 0.001 |
| DFLNnH, LeX/LeX-10 | – | – | – | – | – | – | – | 0.008 | 0.025 | – | 0.12 |
| LeX-Gal (FucLNnT)-07 | – | – | – | – | – | – | – | 0.011 | 0.15 | 240 | 0.012 |
| LeX-Lac (monomeric, LNFIII) | – | – | – | – | – | – | ND | 0.026 | 0.069 | 0.50 | 0.017 |
| LeA-Lac-18 (LNFP II) | – | – | – | – | – | – | – | – | ND | – | 0.013 |
ND = binding observed but KD,App could not be determined (at the highest concentration tested, the signals were not close to saturation).
Figure 4.
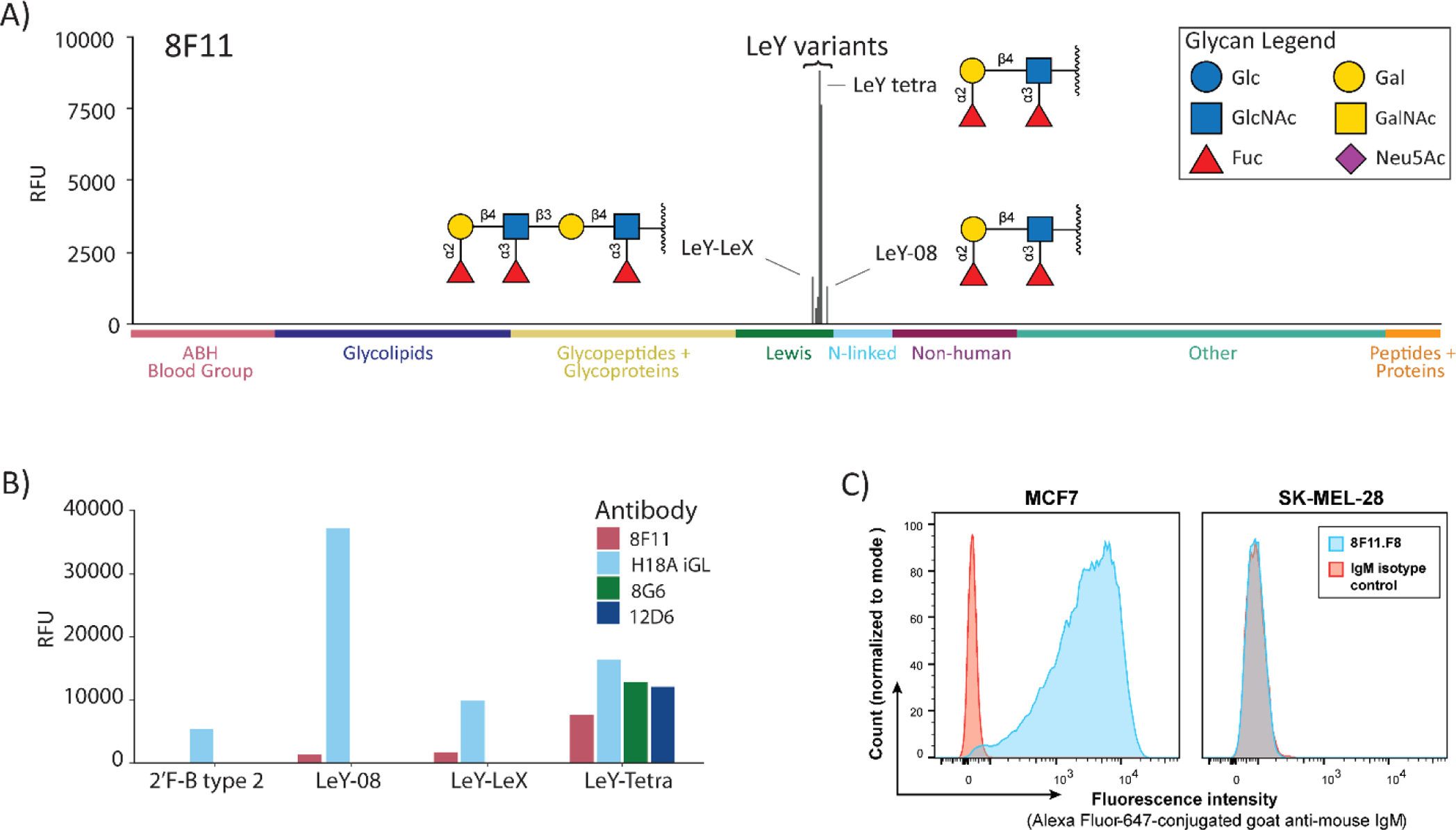
Selective recognition of Lewis Y by germline antibodies. (A) Bar graph showing signals in relative fluorescence units (RFUs) for antibody 8F11 on our 738-component array. Data are shown at a concentration approximately 10-fold higher than the apparent KD value for LeY-tetra to highlight any potential cross-reactivity. Positive and negative controls have been omitted for clarity. Structures are depicted for glycans with signals over 2000 RFU. LeY tetra and LeY-08 have different linkers—see Figure 1. (B) Direct comparison of germline anti-Lewis Y antibody binding to a subset of Lewis Y related glycans. (C) Binding of 8F11 to the Lewis Y positive MCF7 cell line (left) and to the Lewis Y negative SK-MEL-28 cell line (right) as compared to an IgM isotype control. Bar graphs for 12D6 and 8G6 can be found in Figure S3. Additional flow cytometry data can be found in Figures S4 and S5.
In addition to the narrow glycan binding properties, evaluation of potential interactions with a variety of other components on the microarray provided additional evidence that the antibodies were not polyreactive. None of the antibodies bound any of the 16 nonglycosylated peptides or the 45 natural proteins/glycoproteins on the array. These components provide a variety of peptide sequences and protein surfaces for potential recognition by polyreactive antibodies. In addition, the antibodies did not bind our biotin control or any of the linker-only controls on our array. These components display a variety of functional groups such as triazoles, alkynes, alkyl chains, and an ethylene glycol oligomer. Lastly, the antibodies did not bind to DNP-BSA or dsDNA. For comparison, we obtained a polyspecific antibody from another study56 and found that it bound to a range of structurally unrelated molecules on our microarray, including dinitrophenyl-modified BSA, non-glycosylated peptides, and various glycans (see Figure S3B). Thus, binding profiles of polyspecific antibodies on our array are quite distinct from the anti-Lewis Y antibody profiles mentioned above.
In terms of fine specificity for Lewis Y, 8F11, 12D6, and 8G6 displayed key differences depending on the linker or carrier glycan chain at the reducing end of LeY (Figure 4B). Antibody 8F11 bound all three glycans with a terminal Lewis Y (LeY-tetra, LeY-08, and LeY-LeX) but did not recognize glycans with an internal Lewis Y. Antibodies 12D6 and 8G6 only bound LeY-tetra (Figure 4B), a bovine serum albumin conjugate prepared in the same way as the HSA conjugate used for immunizations. Selective binding to LeY-tetra but not LeY-08 or LeY-LeX suggests that the linker region may be involved in recognition. In particular, the amine group in close proximity to the Lewis Y tetrasaccharide could be a key functional group for binding. Carrier chains and linkers have been shown previously to affect antibody recognition through several mechanisms.57–60
High Selectivity is Also Observed in a Cell-Based Assay.
The remarkably high selectivity observed for our germline antibodies was unexpected based on prior studies and the polyspecificity hypothesis. To ensure that this behavior was not unique to the array and to provide further evidence of selectivity, antibodies were tested for cell binding by flow cytometry using MCF7 as a Lewis Y positive cell line and SK-MEL-28 as a Lewis Y negative cell line (Figures 4C, S4, and S5). Antibody 8F11 was able to selectively bind the MCF7 cell line over the SK-MEL-28 cell line (Figure 4C). The ability to recognize a Lewis Y positive cell line but not the myriad of glycans present on the Lewis Y negative cell line further supports high selectivity for this antibody. Interestingly, 12D6 and 8G6 did not bind either cell line (Figure S5), indicating that the fine specificity is important for recognition of Lewis Y on cell surfaces.
A Variety of Other Germline Anti-Glycan Antibodies Display High Selectivity.
To evaluate generality of the AID KO approach and investigate whether high selectivity is a common feature of germline antibodies to carbohydrates, we next immunized AID KO mice with two different immunogens: asialo-ovine submaxillary mucin (aOSM) and inactivated whole MCF7 cells. aOSM is a glycoprotein with over 95% of its carbohydrate content being composed of the tumor-associated Tn antigen (GalNAcα-Ser/Thr).61 MCF7 is a breast cancer cell line. MCF7 cells contain a diverse assortment of carbohydrate antigens and have previously been used as an immunogen to raise monoclonal antibodies to a variety of carbohydrate antigens, including Globo H, blood group H2, Lewis Y, and Lewis A.52,55,62 AID KO mice produced responses to both immunogens, indicating that natural glycoproteins and whole cells are compatible with the AID KO mouse approach.
Next, we carried out fusions to produce hybridoma cells, screening of oligoclonal wells, and subcloning to generate monoclonal antibodies. For oligoclonal wells from an aOSM immunized mouse, supernatants were first screened by ELISA using a 50/50 mixture of Ac-S-Tn(Thr)-A-G-10-BSA and Ac-Tn(S)-Tn(S)-Tn(S)-G-11-BSA. This approach aimed to identify antibodies that recognize single Tn peptides, clustered Tn peptides, or both. Oligoclonal supernatants that were positive for binding via ELISA were then profiled on our microarray. Most (16/18) displayed high selectivity for the Tn antigen, with a strong preference for clustered forms of the Tn antigen. After subcloning, we obtained three clones from the highly selective wells and one from a well that displayed broad binding. Sequencing revealed that all three from the selective wells had the same heavy and light chain sequences, and this antibody was referred to as 9A9. The one from the well with broad reactivity had a distinct sequence, and it was referred to as 8C11 (see Table 1 for germline gene usage and Figure S7 for sequences).
For oligoclonal wells derived from mice immunized with MCF7 cells, we carried out ELISA screening using several mixtures of neoglycoproteins, with the goal of obtaining antibodies with a variety of target specificities. The mixtures contained subsets of neoglycoproteins with the following glycans: Lewis X, Lewis Y, blood group H, sialyl Lewis C, blood group A, maltose oligosaccharides, or the Tn antigen. Most of the 29 oligoclonal wells selected for screening on the microarray displayed high selectivity (Figure S6). After subcloning, we successfully obtained six monoclonal antibodies: 2D3.C11, 4E9, 5B3, 6D5, 11A11, and 16B9. All were determined to be unique sequences (Table 1 and Figure S7). This group included five antibodies to Lewis X and one antibody to sialyl Lewis C. In addition, we observed highly selective anti-glycan binding profiles for two other glycans when screening oligoclonal wells: one well was selective for blood group A and another for blood group H (Figure S6B, wells 15B1, 15D4, and 16A3). Although subcloning was unsuccessful, we can conclude that there was at least one antibody in each well that binds selectively to the associated glycan, providing further evidence for highly specific germline-encoded antibodies that target a variety of carbohydrate antigens.
Affinity and selectivity of the eight germline monoclonal antibodies were evaluated using our glycan microarrays and cell-based assays. Overall, seven of eight showed high selectivity for their primary glycan target in relation to the other glycans on our microarray (Figure 5A, Table 2, and Figure S8). All of the anti-Lewis X antibodies bound best to LeX (dimeric), a glycan with the sequence Galβ1–4(Fucα1–3)GlcNAcβ1–3Galβ1–4(Fucα1–3)GlcNAc (Figure 6A). Intermediate binding was observed to LeX-Gal [Galβ1–4(Fucα1–3)GlcNAcβ1–3Gal] and LeX-Lac [Galβ1–4(Fucα1–3)-GlcNAcβ1–3Galβ1–4Glc], while none bound the simple trisaccharide [Galβ1–4(Fucα1–3)GlcNAcβ1; “LeX trisaccharide”]. Thus, these antibodies were not just selective for Lewis X over other glycans on the array; they also differentiated based on the carrier glycan chain to which Lewis X was attached. Antibody 2D3.C11 had modest affinity but displayed good selectivity for sialyl Lewis C. Antibody 9A9 displayed remarkable selectivity for clustered forms of the Tn antigen, glycopeptides with two or more GalNAcα-Ser/Thr residues in close proximity such as the one shown in Figure 6. The eighth antibody, 8C11, bound most glycans with a terminal GalNAc-α monosaccharide residue, including the Tn antigen, the Forssman antigen, GalNAcα1–3Gal, GalNAcα-Tyr, and blood group A. Although one might characterize 8C11 as selective for GalNAc-α monosaccharide, as opposed to being broadly reactive with unrelated structures, the small size of the epitope renders the antibody functionally polyreactive with many glycan determinants. Therefore, we did not classify it as highly selective. None of these antibodies bound DNP-BSA, dsDNA, nonglycosylated control peptides, proteins, or glycoproteins that do not display the target glycan, biotin, or various linker-only controls. Moreover, five of the seven selective antibodies also bound to antigen positive but not negative cell lines (see Figures 5B,C and S9). The other two, 2D3.C11 and 4E9, did not bind either cell line, presumably due to the weaker affinity of these antibodies (binding to array components was observed but was too weak to determine apparent KD values). These results provide further evidence that the antibodies are highly selective and not polyreactive.
Figure 5.
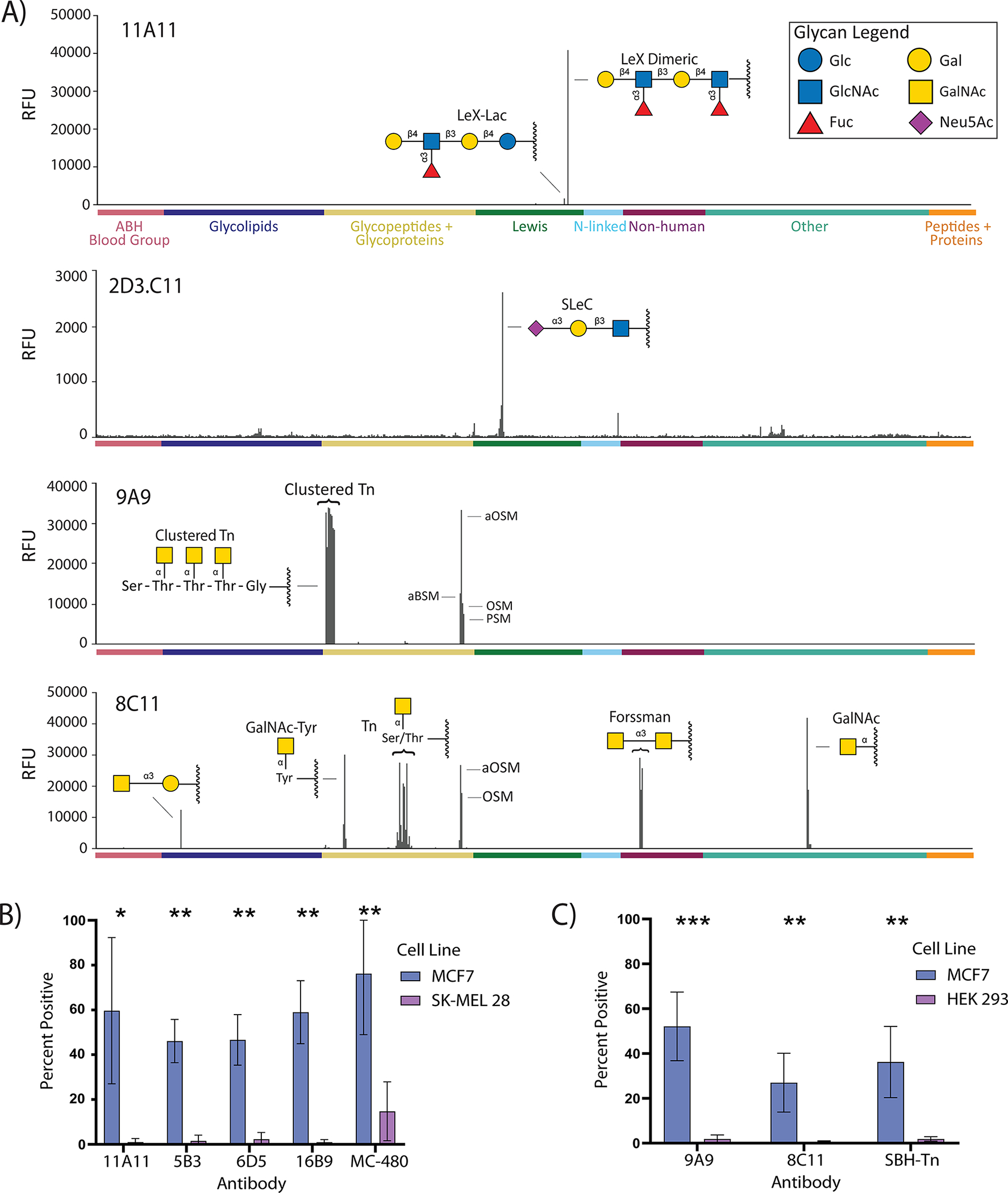
Binding properties of representative germline antibodies to Lewis X, sialyl Lewis C, and the Tn antigen. (A) Signals in relative fluorescence units (RFUs) for antibodies on our 738-component array. Data for 11A11, 9A9, and 8C11 are at a concentration approximately 20-fold higher than KD,App for the primary antigen; 2D3.C11 is at 8 μg/mL. Positive and negative controls have been excluded. (B) Binding data for anti-Lewis X antibodies to Lewis X positive MCF7 cell line and Lewis X negative SK-MEL-28 as compared to positive control anti-Lewis X antibody MC-480. (C) Binding data for anti-Tn antibodies to Tn positive MCF7 cell line and Tn negative HEK-293 cell line as compared to positive control anti-Tn antibody SBH-Tn. Error bars (B,C) represent the SD of three experiments. Additional bar graphs can be found in Figure S8. Representative histograms and additional flow cytometry data can be found in Figure S9.
Figure 6.

Structures of Lewis X (A), sialyl Lewis C (B), and Tn-related (C) array components.
Inferred Germline Antibodies to Lewis Y and the Tn Antigen Display High Selectivity.
Given the unexpectedly high selectivity of the antibodies isolated from AID KO mice, we decided to evaluate two inferred germline antibodies for comparison. The goal was to determine if high selectivity was specific to the AID KO mouse approach or if traditional approaches would also provide highly selective antibodies to these glycans. Because the traditional approach relies on inferring the original germline sequence based on homology to germline genes, we selected two inferred germline antibodies that had the least ambiguity, one derived from the affinity-matured anti-Lewis Y antibody H18A (H18A-iGL) and another derived from the anti-Tn antibody 83D4 (83D4-iGL).63 Because H18A-iGL and 8F11 differed only by two adjacent amino acids located in CDRH3 (RD vs KY; Figure 3B), the comparison would also allow us to also evaluate differences in selectivity for two closely related potential germline antibodies for H18A.
Antibodies were recombinantly expressed as IgGs, purified, and evaluated on our full microarray at several concentrations. The IgG version of 8F11 displayed high selectivity for Lewis Y and an apparent KD value of 460 nM (Figure S10). The large decrease in apparent affinity was expected due to the decreased avidity of an IgG with two binding sites versus an IgM with 10 binding sites. For comparison, H18A-iGL had an apparent KD value of 142 nM for LeY-08, while H18A had an apparent KD value of 21 nM for LeY-08 (Figure S10). Similar to the germline antibodies isolated from the AID KO mice, H18A-iGL demonstrated high selectivity for Lewis Y (Figures S10 and S11). However, there were some key differences in fine specificity (Figure 4B). In particular, H18A-iGL bound best to LeY-08, whereas 8F11 bound LeY-tetra about 25-fold tighter than LeY-08. In addition, H18A-iGL bound to a glycan with an internal Lewis Y (2′F B type 2), whereas 8F11 did not bind this glycan at all. Thus, the inferred germline of H18A and 8F11 has distinct binding properties. These results also demonstrate that positions 108 and/or 109 within the CDRH3 can influence selectivity. Antibody 83D4-iGL displayed high selectivity for clustered Tn peptides (peptides containing two or more Tn residues in close proximity; see Figure S11). Overall, both the AID KO approach and the inferred germline approach yielded anti-glycan antibodies with high selectivity.
Molecular Dynamics and Structural Analysis.
To gain structural insights into recognition by germline anti-glycan antibodies, we employed molecular dynamics simulations. Because germline antibodies 8F11 and 12D6 were highly similar in sequence to several anti-Lewis Y antibodies that had been crystallized, we focused on these. We first built homology models based on the crystal structure of the humanized Fab 3S193 bound to Lewis Y and performed molecular dynamics simulations both with and without bound Lewis Y.64 The simulations revealed that the mature H18A antibody was on average slightly less flexible than the two germline antibodies, in both the bound and unbound state (Figures 7 and S12). Interestingly, upon antigen binding, the CDRH3 loop flexibility in all three antibodies was restrained to a similar level (Figure S12C).
Figure 7.

Modeled structures of antibodies 8F11 and H18A. (A–D) Worm plots illustrating the average magnitude of the RMS atomic fluctuation in each backbone atom over the course of the simulation mapped to the radius of the backbone trace. Regions of the structure that are relatively stable and rigid appear narrower, and more flexible regions of the structure appear wider. View looking down into the binding pocket with the heavy chain on the left and the light chain on the right. CDR loops are labeled, and residues that differ between germline and affinity mature are shown in stick representation.
We also analyzed the conformational space explored by Lewis Y via calculating the RMS displacement for each constituent monosaccharide over time. These distances were binned and normalized, and the resulting histograms are shown in Figure S13. Narrow peaks in the histogram indicate that the antigen is stable and relatively immobile in the binding site, as seen for 8F11 (Figure S13A). In contrast, Lewis Y is unstable within the binding site of 12D6, which is indicated by the constituent monosaccharides moving well away (over 4 Å) from their starting positions (Figure S13B). Interestingly, in H18A, while the galactose and one fucose remain in place, the GlcNAc residue and the other fucose show an increased range of motion (Figure S13C).
To analyze the molecular interactions occurring in the binding pocket, we compared the hydrogen-bonding interactions (including water-bridged interactions) between the crystal structure of 3S193 and those observed at least 10% of the time in the Lewis Y-bound simulations (Figure 8A–D).64 The germline antibody 8F11 retains nearly all the hydrogen bonds seen in the crystal, including tight hydrogen bonds between Asp 109 in the CDRH3 and the GlcNAc residue (Figure 8C,E). For H18A, the hydrogen-bonding interactions are the same, except for the Asp 109–GlcNAc interaction. Residue 109 is a tyrosine in H18A (Figure 8F), which interacts with the GlcNAc residue via a van der Waals stacking interaction instead of hydrogen bonding. Tyrosine and other aromatic residues are known to interact with carbohydrates via CH-π interactions,65 which could explain why we do not observe a loss of affinity associated with this substitution. In the CDRH2, both 8F11 and H18A have Tyr at position 38, which stabilizes the conformation of the loop by forming one wall of the binding site and packing extensively against the GlcNAc ring of Lewis Y. Nearby, Tyr 40 contributes to the network of hydrogen bonds in the binding site (Figure 8A,C–F). Germline antibody 12D6 does not possess tyrosine residues at these positions, which drastically increases flexibility (Figure 9). It can assume a folded-in conformation, where it forms hydrogen bonds to the glycan GlcNAc ring, but it can also fold open. In the latter case, the CDRH2 loop moves outward, such that its sidechains no longer interact with Lewis Y causing the glycan to partially unbind.
Figure 8.

Interactions between Lewis Y and antibodies. (A–D) Schematic view of the hydrogen bond networks. (A) H-bonding for hu3S193 was determined from the crystal structure64; (B–D) H-bonding shown was present at least 10% of the time in the molecular dynamics simulations. (E,F) Cluster representative snapshots illustrating residue differences between antibodies. (E) 8F11 germline, with residue D109 in the CDRH3 loop colored green. (F) H18A affinity mature, with residue Y109 colored green. Hydrogen bonds are shown as orange lines.
Figure 9.

Worm plots, snapshot, and electrostatic surface potential of 12D6. (A,B) Worm plots illustrating the average magnitude of the RMS atomic fluctuation in each backbone atom over the course of the simulation mapped to the radius of the backbone trace. Regions of the structure that are relatively stable and rigid appear narrower, and more flexible regions of the structure appear wider. View looking down into the binding pocket with the heavy chain on the left and the light chain on the right. CDR loops are labeled, and residues that differ between germline and affinity mature are shown in stick representation. (C) Cluster representative snapshot. Hydrogen bonds are shown as orange lines. (D) Top-down view of the antigen binding site surface in 12D6, colored by electrostatic potential. Lewis Y is shown in the pocket, with the linker attachment point marked with a violet ball. The linker is in close proximity to residues D109 and Y110 of the heavy chain (IMGT numbering).
The results from the molecular dynamics simulations, together with the antibody binding results from the glycan microarray, suggest that the linker region of Lewis Y-tetra may be particularly critical for recognition by 12D6. To explore this further, we added the linker to Lewis Y in each of the cluster representative structures from the 12D6 simulation and performed molecular docking calculations using AutoDock. As shown in Figures 9D and S14, the linker docks into the binding pocket in close proximity to the CDRH3. This supports the possibility that within the CDRH3 of 12D6, Asp 109 could be forming a salt-bridged interaction with the positively charged amino group of the Lewis Y-tetra linker, similar to the interaction between Asp 109 and the nonate ester derivative of Lewis Y observed in the crystal structure of BR96.66 The tyrosine present at this position in H18A and H18A-iGL is unlikely to be forming this same salt-bridged interaction, which could explain the difference in linker preference observed for H18A-iGL (Figure 4B). Therefore, variations at CDRH3 position 109 could play a key role in recognition of the carrier glycan or linker.
DISCUSSION
Studies on binding properties of germline antibodies are useful for designing immunogens for vaccines, generating monoclonal antibodies, and understanding antibody development within the immune system. A prime example comes from the HIV field, where germline antibody studies14–17 have enabled novel design strategies to produce an HIV vaccine.67–69 Unfortunately, very little is known about most germline antibodies due to the challenges of obtaining them. Our knowledge of germline antibodies to carbohydrates is especially limited, as anti-carbohydrate antibodies have been understudied and only a small number of monoclonal antibodies to carbohydrates are available.70
In this study, we investigated a unique approach to obtain germline antibodies involving immunization of AID knockout mice. The lack of a functioning AID enzyme results in the absence of somatic hypermutation and isotype class switching,31 and immunization provides an avenue to amplify potentially rare B cells that target a particular antigen of interest. By immunizing with carbohydrate–hapten conjugates, glycoproteins, and whole cells, we successfully generated germline antibodies to a variety of clinically relevant carbohydrate antigens. These included antibodies to tumor-associated carbohydrate antigens (Lewis Y and Tn) and a stage-specific embryonic antigen (Lewis X, also known as SSEA-1 or CD15). Antibodies to these glycans are of special interest for several reasons: antibodies to Lewis X are used in clinical diagnostic assays, CAR T cells incorporating anti-Lewis Y antibodies are currently in several active clinical trials, and CAR T cells incorporating anti-Tn antibodies have shown promising efficacy in mouse models.71
The results provide important insights regarding germline antibody recognition properties. Numerous prior studies have found that germline antibodies to proteins, peptides, small molecule haptens, and bacterial polysaccharides are polyreactive.4–9,12,18,72–75 Some have postulated that selective germline antibodies may also be produced by the immune system, but direct evidence for these has been difficult to obtain. Using the AID knockout mouse approach, we found that most germline antibodies to a variety of glycans, including Lewis Y, Lewis X, Sialyl Lewis C, blood group A, blood group B, blood group H, and the Tn antigen, were very selective. This result builds on our prior finding that inferred germline antibodies to the mammalian glycan GD2 are highly selective for their glycan target.24 In this study, however, the AID KO mouse approach allowed us to isolate germline antibodies to a carbohydrate using a method that does not rely on inferring the germline sequence. Overall, both approaches lead to the conclusion that a variety of glycan-binding germline antibodies are highly selective.
Our results also have important implications for understanding antibody–carbohydrate interactions. A glycan epitope in its natural context is typically attached to the nonreducing end of a glycan chain (acceptor glycan chain), which itself is attached to a protein or lipid at its reducing end. For example, there are at least eight different ways in which Lewis Y can be connected to an acceptor glycan chain in mammals (LeYβ1–2Galβ, β1–3Galβ, β1–4Galβ, β1–6Galβ, β1–2Manα, β1–4Manα, β1–6Manα, and β1–4Manβ).76 In some cases, the acceptor glycan chain can enhance binding by providing additional contacts with the antibody, while in other cases, they are neutral or detrimental.57–60 For most anti-glycan antibodies, little is known about the influence of the acceptor glycan chain. In the cases that have been studied, only affinity-matured antibodies were evaluated. Thus, it was unclear if interactions with the acceptor glycan chain emerge during the affinity-maturation process or if they are present in the germline antibodies. Our data demonstrate that germline antibodies can have distinct preferences based on the acceptor glycan chain. For example, 8F11 recognized Lewis Y when presented on a Lewis X chain (LeY-LeX), while 12D6 and 8G6 did not. The difference in acceptor glycan chain recognition appears to be relevant for binding Lewis Y on cancer cells because only antibody 8F11 bound well to the Lewis Y positive cell line. The acceptor glycan chain also affected recognition for anti-Lewis X antibodies.
The ability of germline antibodies and the corresponding naïve B cells to recognize the acceptor glycan chain is an important consideration when designing carbohydrate-based immunogens. Small carbohydrates are not immunogenic on their own and are typically conjugated to a carrier protein when generating an immunogen.77 Alternatively, one may use a natural glycoprotein or cells that display the target glycan as an immunogen. These variations can lead to different displays of the glycan antigen. For example, cells may display Lewis Y attached to Galβ1–4Glcβ-ceramide as part of the cell membrane, whereas the Lewis Y tetrasaccharide might be attached to a Galβ1–3GalNAcα-serine when displayed on a glycoprotein. Alternatively, it might be connected via an unnatural, synthetic linker to a carrier protein such as CRM197. While all three display Lewis Y to the immune system, the structures of the carrier molecules are different. Thus, they could activate different naïve B cells and, subsequently, produce different pools of affinity-matured antibodies with distinct selectivities. At present, however, little is known about the effects of different carrier molecules or how best to design the carrier molecule to generate antibodies with the desired properties. Additional studies in this area would be useful. Also, increasing the diversity of acceptor glycan chains on carbohydrate microarrays will be worthwhile to better understand when and how they influence recognition. Advances in chemoenzymatic synthesis of glycans could be especially useful for this objective.78
Several limitations of our study should be noted. First, although our glycan array includes a large and diverse collection of glycans and other components, it contains only a small fraction of the glycan diversity found in nature. To help address this limitation, we evaluated binding to antigen positive and negative cell lines, which display a broad assortment of other glycans as well as nonglycan antigens. The cell binding results were consistent with high selectivity; however, it is possible that the antibodies in our study bind other antigens that were not present on our array or the negative cell lines. Second, despite evaluating a diverse panel of germline antibodies, herein, this group represents only a small sample of all anti-glycan antibodies found within naïve antibody repertoires. Moreover, the set of antibodies obtained in a study like this may depend on the type of immunogens, immunization protocols, adjuvants, screening protocol, and/or mouse strains. We have partly addressed the issue of generality by (a) immunizing with three different types of antigens (a carbohydrate protein conjugate, a natural glycoprotein, and whole cells), (b) assessing properties of antibodies toward multiple carbohydrate targets, and (c) evaluating both inferred germlines and germline antibodies isolated from AID knockout mice. Nevertheless, additional studies will be needed to investigate our findings more fully. Third, although the AID KO approach provides access to germline antibodies, it does not provide affinity-matured antibodies from those germline antibodies. Therefore, one does not get matched pairs of germline and affinity-matured antibodies to evaluate the immunological evolution process. Consequently, we consider it to be a complementary approach to the inferred germline approach, rather than a replacement.
Lastly, we note several broader implications of our results. Inducing antibodies to carbohydrates is notoriously difficult, especially for certain classes of glycans such as tumor-associated carbohydrate antigens.70 High selectivity of the associated naïve B cell receptors could be an important contributing factor. In these situations, the structure of the immunogen must be precise to activate the associated naïve B cells and initiate an antibody response; thus, there is very little room for error when designing or selecting immunogens. Our results also raise questions about why certain germline antibodies/receptors have high selectivity and the mechanisms by which the immune system produces highly selective germline antibodies. Avoiding autoimmunity could be a key factor. Many carbohydrate antigens for which an antibody response would be beneficial are very similar in structure to other mammalian glycans, sometimes only differing by the stereochemistry of a single hydroxyl group. Therefore, even modest levels of polyreactivity could result in significant reaction with self-glycans. To avoid this problem, certain V, D, and J genes that give rise to antibodies that bind broadly to glycans may have been eliminated through years of evolution and natural selection. Alternatively, B cells that react broadly with glycans may be generated within the body but then removed via mechanisms that actively eradicate autoreactive immature B cells from the bone marrow or new emigrants from the periphery.30,79 Finally, the existence of both polyreactive and highly selective germline antibodies indicates two distinct pathways for antibody development: polyreactive germline antibodies provide broad reactivity for antigen families that are very distinct from mammalian structures, and highly selective germline antibodies provide careful targeting for self-like antigens where autoimmune reactivity is a potential problem. Based on this model, there are likely to be many other highly selective germline antibodies that have not yet been characterized.
MATERIALS AND METHODS
Immunizations and Production of Hybridomas.
AID−/− C57BL/6 mice were obtained from Riken BRC (Japan; cat# RBRC06299), deposited by Honjo.31 All mouse work was carried out by Green Mountain Antibodies, Inc. (Burlington, VT). Animal protocols and works were approved by the Institutional Animal Care and Use Committee of Green Mountain Antibodies, Inc. [OLAW Assurance ID is D16–00647 (A4289–01); protocol #LewisY-A.01: 1788, aOSM-A.01: 1944, and MCF7-A.01: 1932]. The human breast cancer cell line MCF7 was generously provided by the NCI Developmental Therapeutics Program and was authenticated by ATCC’s cell line authentication service (ATCC 135-XV).
Full details for the immunization can be found in the Supporting Information. Briefly, three female mice were immunized with the Lewis Y-human serum albumin conjugate (for preparation of discussed conjugates, see Supporting Information Methods and Figures S15 and S16) in complete Freund’s adjuvant and boosted several times. Due to low titers (~1:8000), these mice were boosted with LeY-tetra-HSA in the Sigma Adjuvant System (SAS) twice and then with LeY-tetra-HSA in Titermax adjuvant twice more. The mouse with the highest titers (1:24,000) was boosted a final time, followed by harvesting the spleen and hybridoma generation. ELISA screening was carried out using LeY-tetra-BSA.
Additionally, three male mice were immunized with aOSM in complete Freund’s adjuvant followed by three boosts in incomplete Freund’s adjuvant and then two additional boosts in SAS adjuvant. ELISA screening was carried out using Ac-S-Tn(Thr)-A-G.80,81 The mouse with the highest titers was boosted a final time with aOSM in PBS, followed by spleen harvesting and hybridoma fusion. A final cohort of three female mice was immunized with whole MCF7 cells in PBS treated with 5 μg/mL Cytochalasin B and 0.02% azide to inactivate. Mice were boosted four times with MCF7 cells in PBS, followed by spleen harvesting and hybridoma fusion. Because the goal with MCF7-immunized mice was to isolate germline antibodies to a variety of different glycans, we used a modified screening protocol for the hybridomas. At the oligoclonal stage, wells were screened by ELISA for binding to two glycan antigen mixtures (antigen mix 1 = Lewis X, Lewis Y, and blood group H; antigen mix 2 = sialyl Lewis C, blood group A, and the Tn antigen). Hybridoma supernatants from 28 positive wells were then profiled individually on our glycan microarray. After screening, six positive wells were successfully subcloned.
Glycan Microarray Fabrication, Assay, and Data Analysis.
Microarray fabrication, assay, and data analysis were carried out as previously reported (see also Supporting Information).48,82 Screening and analysis of selectivity was carried out using an array with 738 array components (A738) or 816 array components (A816) printed in 8 replicate blocks. The array slide was covered with a ProPlate 8-well chamber (Grace Bio-labs, Bend, OR), and each well was blocked with PBS + 3% w/v BSA. Slides were washed several times with PBST (PBS + 0.05% v/v Tween 20), and then, mouse sera, hybridoma supernatants, or purified antibodies were added at various dilutions in PBST + 1% w/v BSA. The purified antibodies tested were the inferred germline of H18A (H18A-iGL), 8F11, and 83D4-iGL, each of which was recombinantly expressed as IgGs (for experimental details and sequences of these antibodies, see Supporting Information Methods and Figures S17–S21). Wells were probed with fluorophore-conjugated secondary antibodies in PBS + 1% BSA. Arrays were again washed with PBST, the eight-well chamber was removed, and the slide was submerged in PBST. Slides were dried by centrifugation prior to scanning. To determine apparent KD values, hybridoma supernatants were serially diluted in PBST + 1% BSA and then assayed as mentioned above with smaller, 16-well focused microarrays. The concentration of IgM in each supernatant was determined using a mouse IgM ELISA kit following the manufacturer’s instructions (Innovative Research; see Table S1).
Scanning was conducted using an InnoScan 1100 AL fluorescence scanner (Innopsys; Chicago, IL). Analysis was performed using GenePix Pro 7 software (Molecular Devices Corporation; Sunnyvale, CA). Missing spots on the array (determined by a scan of the slide prior to the experiment) were flagged and excluded from analysis. Background fluorescence was subtracted from the median fluorescence, and values were averaged for duplicate spots for the full array or averaged over four to six spots for the apparent KD experiments (two spots per array experiment, performed in duplicate or triplicate). The Supporting Information Excel File provides the full microarray data, and Figure S22 shows the representative array images. Apparent dissociation constants (KD,App) were calculated using the GraphPad Prism software following the method of MacBeath (see also Table 2, Supporting Information Methods, and Figures S2 and S10).83
Cell Staining, Fixation, and Flow Cytometry.
The human breast cancer cell line MCF7 and the human melanoma cell line SK-MEL-28 were generously provided by the NCI Developmental Therapeutics Program. The human embryonic kidney HEK-293 cell line was purchased from the American Type Culture Collection (ATCC CRL-1573; Manassas, VA). All cells were authenticated by ATCC’s cell line authentication service (ATCC 135-XV) and grown in either DMEM or RPMI-1640 media supplemented with 10% FBS, 1% Pen-Strep, and 2 mM GlutaMAX at 37 °C and 5% CO2.
Cells were dissociated with 0.25% trypsin–EDTA (Thermo Fisher Scientific, Inc) or TrypLE Express Enzyme (Thermo Fisher Scientific, Inc.). Between 0.7 and 2.5 million cells were divided into each condition. MCF7 and HEK-293 cell lines were used to evaluate anti-Tn and anti-SLeC antibodies, SK-MEL-28 and HEK-293 were used to evaluate anti-LeX antibodies, and MCF7 and SK-MEL 28 cells were used to evaluate anti-LeY antibodies. Cells were blocked with PBS with 3% w/v BSA. Cells were then incubated with the hybridoma supernatant, anti-glycan control antibodies (2D3: TCI America A2509, MC480: BioLegend 125602, SBH-TN: SBH Sciences 041212), or mouse IgM isotype control (Sigma M5909; Invitrogen 02–6800). Cells receiving a primary were then probed with an Alexa Fluor 647-conjugated Goat Anti-Mouse IgM antibody (Jackson 115–605-075) in PBS with 1% w/v BSA. All cells were fixed with 4% PFA. Flow cytometry analyses were conducted on a BD LSR II flow cytometer equipped with a 635 nm red diode laser. All experiments were run in duplicate or triplicate, apart from the 8G6, 12D6, 4E9, and 2D3.C11 conditions, which were each conducted once (see also Figures S4, S5, and S9).
Molecular Dynamics.
A full description of the molecular modeling methods is available in the Supporting Information. Briefly, homology models of 8F11, 12D6, and the mature antibody H18A were built based on the crystal structure of hu3S193.64 Multicopy molecular dynamics simulations were run for the three antibodies, with and without bound Lewis Y, for a total simulation time of 2 μs for each of the six systems. The interactions of the linker between the glycan and the carrier protein were explored using covalent docking.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Consortium for Functional Glycomics (GM62116; The Scripps Research Institute), Xuefei Huang (Michigan State University), Thomas Tolbert (University of Kansas), Lai-Xi Wang (University of Maryland), Joseph Barchi, Jr. (National Cancer Institute), Todd Lowary (University of Alberta), Omicron Biochemicals Inc., GlycoHub, and Glycan Therapeutics for generously contributing glycans for the array. We thank the NCI Division of Cancer Treatment and Diagnosis (Developmental Therapeutics Program, NCI-60 Human Tumor Cell Lines Screen) for generously providing cell lines. We thank the CCR-Frederick Flow Cytometry Core for assistance with flow cytometry experiments. We thank Green Mountain Antibodies, Inc., for helpful discussions and for their contributions to the sequencing of the antibodies. The molecular dynamics simulations utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). This work was supported in part by the Intramural Research Program of the National Cancer Institute, NIH and in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.1c12745.
Contributor Information
Andrew T. DeLaitsch, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States.
Jacey R. Pridgen, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States.
Avery Tytla, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States.
Megan L. Peach, Basic Science Program, Chemical Biology Laboratory, Leidos Biomedical Inc., Frederick National Laboratory for Cancer Research, Frederick, Maryland 21702, United States
Rayleen Hu, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States.
David W. Farnsworth, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States
Aislinn K. McMillan, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States
Natalie Flanagan, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States.
J. Sebastian Temme, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States.
Marc C. Nicklaus, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States
Jeffrey C. Gildersleeve, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States
REFERENCES
- (1).Schroeder HW Jr.; Cavacini L Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010, 125, S41–S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Leavy O Therapeutic antibodies: Past, present and future. Nat. Rev. Immunol. 2010, 10, 297. [DOI] [PubMed] [Google Scholar]
- (3).Vajda S; Porter KA; Kozakov D Progress toward improved understanding of antibody maturation. Curr. Opin. Struct. Biol. 2021, 67, 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Jimenez R; Salazar G; Yin J; Joo T; Romesberg FE Protein dynamics and the immunological evolution of molecular recognition. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 3803–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Nguyen HP; Seto NOL; MacKenzie CR; Brade L; Kosma P; Brade H; Evans SV Germline antibody recognition of distinct carbohydrate epitopes. Nat. Struct. Biol. 2003, 10, 1019–1025. [DOI] [PubMed] [Google Scholar]
- (6).Patten PA; Gray NS; Yang PL; Marks CB; Wedemayer GJ; Boniface JJ; Stevens RC; Schultz PG The immunological evolution of catalysis. Science 1996, 271, 1086–1091. [DOI] [PubMed] [Google Scholar]
- (7).Wedemayer GJ; Patten PA; Wang LH; Schultz PG; Stevens RC Structural insights into the evolution of an antibody combining site. Science 1997, 276, 1665–1669. [DOI] [PubMed] [Google Scholar]
- (8).Zimmermann J; Oakman EL; Thorpe IF; Shi X; Abbyad P; Brooks CL Iii; Boxer SG; Romesberg FE Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 13722–13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Manivel V; Sahoo NC; Salunke DM; Rao KVS Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity 2000, 13, 611–620. [DOI] [PubMed] [Google Scholar]
- (10).Sethi DK; Agarwal A; Manivel V; Rao KVS; Salunke DM Differential Epitope Positioning within the Germline Antibody Paratope Enhances Promiscuity in the Primary Immune Response. Immunity 2006, 24, 429–438. [DOI] [PubMed] [Google Scholar]
- (11).Adhikary R; Yu W; Oda M; Walker RC; Chen T; Stanfield RL; Wilson IA; Zimmermann J; Romesberg FE Adaptive mutations alter antibody structure and dynamics during affinity maturation. Biochemistry 2015, 54, 2085–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Romesberg FE; Spiller B; Schultz PG; Stevens RC Immunological origins of binding and catalysis in a Diels-Alderase antibody. Science 1998, 279, 1929–1933. [DOI] [PubMed] [Google Scholar]
- (13).Yin J; Andryski SE; Beuscher AE; Stevens RC; Schultz PG Structural evidence for substrate strain in antibody catalysis. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hoot S; McGuire AT; Cohen KW; Strong RK; Hangartner L; Klein F; Diskin R; Scheid JF; Sather DN; Burton DR; Stamatatos L Recombinant HIV Envelope Proteins Fail to Engage Germline Versions of Anti-CD4bs bNAbs. PLoS Pathog. 2013, 9, No. e1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Scharf L; West AP Jr.; Gao H; Lee T; Scheid JF; Nussenzweig MC; Bjorkman PJ; Diskin R Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Scharf L; West AP Jr.; Sievers SA; Chen C; Jiang S; Gao H; Gray MD; McGuire AT; Scheid JF; Nussenzweig MC; Stamatatos L; Bjorkman PJ Structural basis for germline antibody recognition of HIV-1 immunogens. eLife 2016, 5, No. e13783.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Xiao X; Chen W; Feng Y; Zhu Z; Prabakaran P; Wang Y; Zhang M-Y; Longo NS; Dimitrov DS Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: Implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 2009, 390, 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Evans DW; Muller-Loennies S; Brooks CL; Brade L; Kosma P; Brade H; Evans SV Structural insights into parallel strategies for germline antibody recognition of lipopolysaccharide from Chlamydia. Glycobiology 2011, 21, 1049–1059. [DOI] [PubMed] [Google Scholar]
- (19).Willis JR; Briney BS; DeLuca SL; Crowe JE Jr.; Meiler J Human Germline Antibody Gene Segments Encode Polyspecific Antibodies. PLoS Comput. Biol. 2013, 9, No. e1003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Schmidt AG; Xu H; Khan AR; O’Donnell T; Khurana S; King LR; Manischewitz J; Golding H; Suphaphiphat P; Carfi A; Settembre EC; Dormitzer PR; Kepler TB; Zhang R; Moody MA; Haynes BF; Liao H-X; Shaw DE; Harrison SC Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Fernández-Quintero ML; Loeffler JR; Bacher LM; Waibl F; Seidler CA; Liedl KR Local and Global Rigidification Upon Antibody Affinity Maturation. Front. Mol. Biosci. 2020, 7, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Thorpe IF; Brooks CL III Molecular evolution of affinity and flexibility in the immune system. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Babor M; Kortemme T Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility. Proteins 2009, 75, 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Sterner E; Peach ML; Nicklaus MC; Gildersleeve JC Therapeutic Antibodies to Ganglioside GD2 Evolved from Highly Selective Germline Antibodies. Cell Rep. 2017, 20, 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ye J; Ma N; Madden TL; Ostell JM IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013, 41, W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Brochet X; Lefranc M-P; Giudicelli V IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008, 36, W503–W508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Munshaw S; Kepler TB SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements. Bioinformatics 2010, 26, 867–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Tsuiji M; Yurasov S; Velinzon K; Thomas S; Nussenzweig MC; Wardemann H A checkpoint for autoreactivity in human IgM + memory B cell development. J. Exp. Med. 2006, 203, 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zikherman J; Parameswaran R; Weiss A Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature 2012, 489, 160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wardemann H; Yurasov S; Schaefer A; Young JW; Meffre E; Nussenzweig MC Predominant autoantibody production by early human B cell precursors. Science 2003, 301, 1374–1377. [DOI] [PubMed] [Google Scholar]
- (31).Muramatsu M; Kinoshita K; Fagarasan S; Yamada S; Shinkai Y; Honjo T Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 2000, 102, 553–563. [DOI] [PubMed] [Google Scholar]
- (32).Noble P; Spendlove I; Harding S; Parsons T; Durrant LG Therapeutic Targeting of Lewisy and Lewisb with a Novel Monoclonal Antibody 692/29. PLoS One 2013, 8, No. e54892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Herbertson RA; Tebbutt NC; Lee F-T; MacFarlane DJ; Chappell B; Micallef N; Lee S-T; Saunder T; Hopkins W; Smyth FE; Wyld DK; Bellen J; Sonnichsen DS; Brechbiel MW; Murone C; Scott AM Phase I biodistribution and pharmacokinetic study of Lewis Y-targeting immunoconjugate CMD-193 in patients with advanced epithelial cancers. Clin. Cancer Res. 2009, 15, 6709–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kelly MP; Lee FT; Smyth FE; Brechbiel MW; Scott AM Enhanced efficacy of 90Y-radiolabeled anti-lewis Y humanized monoclonal antibody hu3S193 and paclitaxel combined-modality radioimmunotherapy in a breast cancer model. J. Nucl. Med. 2006, 47, 716–725. [PubMed] [Google Scholar]
- (35).Boghaert ER; Sridharan L; Armellino DC; Khandke KM; DiJoseph JF; Kunz A; Dougher MM; Jiang F; Kalyandrug LB; Hamann PR; Frost P; Damle NK Antibody-targeted chemotherapy with the calicheamicin conjugate hu3S193-N-Acetyl γ calicheamicin dimethyl hydrazide targets lewis y and eliminates lewisy-positive human carcinoma cells and xenografts. Clin. Cancer Res. 2004, 10, 4538–4549. [DOI] [PubMed] [Google Scholar]
- (36).Posey JA; Khazaeli MB; Bookman MA; Nowrouzi A; Grizzle WE; Thornton J; Carey DE; Lorenz JM; Sing AP; Siegall CB; LoBuglio AF; Saleh MN A phase I trial of the single-chain immunotoxin SGN-10 (BR96 sFv-PE40) in patients with advanced solid tumors. Clin. Cancer Res. 2002, 8, 3092. [PubMed] [Google Scholar]
- (37).Lövqvist A; Humm JL; Sheikh A; Finn RD; Koziorowski J; Ruan S; Pentlow KS; Jungbluth A; Welt S; Lee FT; Brechbiel MW; Larson SM PET imaging of 86Y-labeled anti-Lewis Y monoclonal antibodies in a nude mouse model: Comparison between 86Y and 111In radiolabels. J. Nucl. Med. 2001, 42, 1281. [PubMed] [Google Scholar]
- (38).Scott AM; Geleick D; Rubira M; Clarke K; Nice EC; Smyth FE; Stockert E; Richards EC; Carr FJ; Harris WJ; Armour KL; Rood J; Kypridis A; Kronina V; Murphy R; Lee FT; Liu Z; Kitamura K; Ritter G; Laughton K; Hoffman E; Burgess AW; Old LJ Construction, production, and characterization of humanized anti-Lewis Y monoclonal antibody 3S193 for targeted immunotherapy of solid tumors. Cancer Res. 2000, 60, 3254. [PubMed] [Google Scholar]
- (39).Clarke K; Lee FT; Brechbiel MW; Smyth FE; Old LJ; Scott AM In vivo biodistribution of a humanized anti-Lewis Y monoclonal antibody (hu3s193) in MCF-7 xenografted BALB/c nude mice. Cancer Res. 2000, 60, 4804–4811. [PubMed] [Google Scholar]
- (40).Clarke K; Lee FT; Brechbiel MW; Smyth FE; Old LJ; Scott AM Therapeutic efficacy of anti-Lewis(y) humanized 3S193 radioimmunotherapy in a breast cancer model: Enhanced activity when combined with taxol chemotherapy. Clin. Cancer Res. 2000, 6, 3621–3628. [PubMed] [Google Scholar]
- (41).Myers RB; Srivastava S; Grizzle WE Lewis Y antigen as detected by the monoclonal antibody BR96 is expressed strongly in prostatic adenocarcinoma. J. Urol. 1995, 153, 1572–1574. [PubMed] [Google Scholar]
- (42).Heimburg-Molinaro J; Lum M; Vijay G; Jain M; Almogren A; Rittenhouse-Olson K Cancer vaccines and carbohydrate epitopes. Vaccine 2011, 29, 8802–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hyun JY; Pai J; Shin I The Glycan Microarray Story from Construction to Applications. Acc. Chem. Res. 2017, 50, 1069–1078. [DOI] [PubMed] [Google Scholar]
- (44).Geissner A; Seeberger PH Glycan Arrays: From Basic Biochemical Research to Bioanalytical and Biomedical Applications. Annu. Rev. Anal. Chem. 2016, 9, 223–247. [DOI] [PubMed] [Google Scholar]
- (45).Palma AS; Feizi T; Childs RA; Chai W; Liu Y The neoglycolipid (NGL)-based oligosaccharide microarray system poised to decipher the meta-glycome. Curr. Opin. Chem. Biol. 2014, 18, 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Arthur CM; Cummings RD; Stowell SR Using glycan microarrays to understand immunity. Curr. Opin. Chem. Biol. 2014, 18, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Park S; Gildersleeve JC; Blixt O; Shin I Carbohydrate Microarrays. Chem. Soc. Rev. 2013, 42, 4310–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Xia L; Gildersleeve JC The glycan array platform as a tool to identify carbohydrate antigens. Methods Mol. Biol. 2015, 1331, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Gildersleeve JC Insights into Antibody-Carbohydrate Recognition from Neoglycoprotein Microarrays. ACS Symp. Ser. 2020, 1346, 23–37. [Google Scholar]
- (50).Stowell CP; Lee YC Neoglycoproteins: the preparation and application of synthetic glycoproteins. Adv. Carbohydr. Chem. Biochem. 1980, 37, 225–281. [DOI] [PubMed] [Google Scholar]
- (51).Kaneko T; Iba Y; Zenita K; Shigeta K; Nakano E; Itoh W; Kurosawa Y; Kannagi R; Yasukawa K Preparation of mouse-human chimeric antibody to an embryonic carbohydrate antigen, Lewis Y. J. Biochem. 1993, 113, 114–117. [DOI] [PubMed] [Google Scholar]
- (52).Kitamura K; Stockert E; Garin-Chesa P; Welt S; Lloyd KO; Armour KL; Wallace TP; Harris WJ; Carr FJ; Old LJ Specificity analysis of blood group Lewis-y (Le(y)) antibodies generated against synthetic and natural Le(y) determinants. Proc. Natl. Acad. Sci. U.S.A. 1994, 91, 12957–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Blaszczyk-Thurin M; Thurin J; Hindsgaul O; Karlsson KA; Steplewski Z; Koprowski HY and blood group B type 2 glycolipid antigens accumulate in a human gastric carcinoma cell line as detected by monoclonal antibody. Isolation and characterization by mass spectrometry and NMR spectroscopy. J. Biol. Chem. 1987, 262, 372–379. [PubMed] [Google Scholar]
- (54).Hellström I; Garrigues HJ; Garrigues U; Hellström KE Highly tumor-reactive, internalizing, mouse monoclonal antibodies to Le(y)-related cell surface antigens. Cancer Res. 1990, 50, 2183–2190. [PubMed] [Google Scholar]
- (55).Pastan I; Lovelace ET; Gallo MG; Rutherford AV; Magnani JL; Willingham MC Characterization of monoclonal antibodies B1 and B3 that react with mucinous adenocarcinomas. Cancer Res. 1991, 51, 3781. [PubMed] [Google Scholar]
- (56).Shehata L; Maurer DP; Wec AZ; Lilov A; Champney E; Sun T; Archambault K; Burnina I; Lynaugh H; Zhi X; Xu Y; Walker LM Affinity Maturation Enhances Antibody Specificity but Compromises Conformational Stability. Cell Rep. 2019, 28, 3300–3308. [DOI] [PubMed] [Google Scholar]
- (57).Liu L; Zha J; Digiandomenico A; McAllister D; Stover CK; Wang Q; Boons G-J Synthetic Enterobacterial Common Antigen (ECA) for the Development of a Universal Immunotherapy for Drug-ResistantEnterobacteriaceae. Angew. Chem., Int. Ed. 2015, 54, 10953–10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Johnson MA; Cartmell J; Weisser NE; Woods RJ; Bundle DR Molecular recognition of Candida albicans (1→2)-β-mannan oligosaccharides by a protective monoclonal antibody reveals the immunodominance of internal saccharide residues. J. Biol. Chem. 2012, 287, 18078–18090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Bovin N; Obukhova P; Shilova N; Rapoport E; Popova I; Navakouski M; Unverzagt C; Vuskovic M; Huflejt M Repertoire of human natural anti-glycan immunoglobulins. Do we have auto-antibodies? Biochim. Biophys. Acta, Gen. Subj. 2012, 1820, 1373–1382. [DOI] [PubMed] [Google Scholar]
- (60).Gildersleeve JC; Wright WS Diverse molecular recognition properties of blood group A binding monoclonal antibodies. Glycobiology 2016, 26, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Hill HD; Reynolds JA; Hill RL Purification, composition, molecular weight, and subunit structure of ovine submaxillary mucin. J. Biol. Chem. 1977, 252, 3791–3798. [PubMed] [Google Scholar]
- (62).Mènard S; Tagliabue E; Canevari S; Fossati G; Colnaghi MI Generation of Monoclonal Antibodies Reacting with Normal and Cancer Cells of Human Breast. Cancer Res. 1983, 43, 1295–1300. [PubMed] [Google Scholar]
- (63).Charpin C; Pancino G; Osinaga E; Bonnier P; Lavaut MN; Allasia C; Roseto A Monoclonal antibody 83D4 immunoreactivity in human tissues: Cellular distribution and microcytophotometric analysis of immunoprecipitates on tissue sections. Anticancer Res. 1992, 12, 209–223. [PubMed] [Google Scholar]
- (64).Ramsland PA; Farrugia W; Bradford TM; Mark Hogarth P; Scott AM Structural Convergence of Antibody Binding of Carbohydrate Determinants in Lewis Y Tumor Antigens. J. Mol. Biol. 2004, 340, 809–818. [DOI] [PubMed] [Google Scholar]
- (65).Hudson KL; Bartlett GJ; Diehl RC; Agirre J; Gallagher T; Kiessling LL; Woolfson DN Carbohydrate-Aromatic Interactions in Proteins. J. Am. Chem. Soc. 2015, 137, 15152–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Jeffrey PD; Bajorath J; Chang CY; Yelton D; Hellström I; Hellström KE; Sheriff S The X-ray structure of an anti-tumour antibody in complex with antigen. Nat. Struct. Biol. 1995, 2, 466–471. [DOI] [PubMed] [Google Scholar]
- (67).Tian M; Cheng C; Chen X; Duan H; Cheng H-L; Dao M; Sheng Z; Kimble M; Wang L; Lin S; Schmidt SD; Du Z; Joyce MG; Chen Y; DeKosky BJ; Chen Y; Normandin E; Cantor E; Chen RE; Doria-Rose NA; Zhang Y; Shi W; Kong W-P; Choe M; Henry AR; Laboune F; Georgiev IS; Huang P-Y; Jain S; McGuire AT; Georgeson E; Menis S; Douek DC; Schief WR; Stamatatos L; Kwong PD; Shapiro L; Haynes BF; Mascola JR; Alt FW Induction of HIV Neutralizing Antibody Lineages in Mice with Diverse Precursor Repertoires. Cell 2016, 166, 1471–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Sok D; Briney B; Jardine JG; Kulp DW; Menis S; Pauthner M; Wood A; Lee E-C; Le KM; Jones M; Ramos A; Kalyuzhniy O; Adachi Y; Kubitz M; MacPherson S; Bradley A; Friedrich GA; Schief WR; Burton DR Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 2016, 353, 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Escolano A; Gristick HB; Abernathy ME; Merkenschlager J; Gautam R; Oliveira TY; Pai J; West AP Jr.; Barnes CO; Cohen AA; Wang H; Golijanin J; Yost D; Keeffe JR; Wang Z; Zhao P; Yao K-H; Bauer J; Nogueira L; Gao H; Voll AV; Montefiori DC; Seaman MS; Gazumyan A; Silva M; McGuire AT; Stamatatos L; Irvine DJ; Wells L; Martin MA; Bjorkman PJ; Nussenzweig MC Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Nature 2019, 570, 468–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Sterner E; Flanagan N; Gildersleeve JC Perspectives on Anti-Glycan Antibodies Gleaned from Development of a Community Resource Database. ACS Chem. Biol. 2016, 11, 1773–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Posey AD; Schwab RD; Boesteanu AC; Steentoft C; Mandel U; Engels B; Stone JD; Madsen TD; Schreiber K; Haines KM; Cogdill AP; Chen TJ; Song D; Scholler J; Kranz DM; Feldman MD; Young R; Keith B; Schreiber H; Clausen H; Johnson LA; June CH Engineered CAR T Cells Targeting the Cancer-Associated Tn-Glycoform of the Membrane Mucin MUC1 Control Adenocarcinoma. Immunity 2016, 44, 1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Brooks CL; Müller-Loennies S; Brade L; Kosma P; Hirama T; MacKenzie CR; Brade H; Evans SV. Exploration of Specificity in Germline Monoclonal Antibody Recognition of a Range of Natural and Synthetic Epitopes. J. Mol. Biol. 2008, 377, 450–468. [DOI] [PubMed] [Google Scholar]
- (73).Gerstenbruch S; Brooks CL; Kosma P; Brade L; MacKenzie CR; Evans SV; Brade H; Muller-Loennies S Analysis of cross-reactive and specific anti-carbohydrate antibodies against lipopolysaccharide from Chlamydophila psittaci. Glycobiology 2009, 20, 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Wedemayer GJ; Wang LH; Patten PA; Schultz PG; Stevens RC Crystal structures of the free and liganded form of an esterolytic catalytic antibody. J. Mol. Biol. 1997, 268, 390–400. [DOI] [PubMed] [Google Scholar]
- (75).Wong SE; Sellers BD; Jacobson MP Effects of somatic mutations on CDR loop flexibility during affinity maturation. Proteins: Struct., Funct., Bioinf. 2011, 79, 821–829. [DOI] [PubMed] [Google Scholar]
- (76).Cummings RD The repertoire of glycan determinants in the human glycome. Mol. BioSyst. 2009, 5, 1087–1104. [DOI] [PubMed] [Google Scholar]
- (77).Lang S; Huang X Carbohydrate Conjugates in Vaccine Developments. Front. Chem. 2020, 8, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Na L; Li R; Chen X Recent progress in synthesis of carbohydrates with sugar nucleotide-dependent glycosyltransferases. Curr. Opin. Chem. Biol. 2021, 61, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Wardemann H; Nussenzweig MC B-Cell Self-Tolerance in Humans. Advances in Immunology; Academic Press, 2007; Vol. 95, pp 83–110. [DOI] [PubMed] [Google Scholar]
- (80).Zhang Y; Muthana SM; Farnsworth D; Ludek O; Adams K; Barchi JJ; Gildersleeve JC Enhanced Epimerization of Glycosylated Amino Acids During Solid Phase Peptide Synthesis. J. Am. Chem. Soc. 2012, 134, 6316–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Zhang Y; Muthana SM; Barchi JJ; Gildersleeve JC Divergent Behavior of Glycosylated Threonine and Serine Derivatives in Solid Phase Peptide Synthesis. Org. Lett. 2012, 14, 3958–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Campbell CT; Zhang Y; Gildersleeve JC Construction and Use of Glycan Microarrays. Curr. Protoc. Chem. Biol. 2010, 2, 37–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Gordus A; MacBeath G Circumventing the problems caused by protein diversity in microarrays: Implications for protein interaction networks. J. Am. Chem. Soc. 2006, 128, 13668–13669. [DOI] [PubMed] [Google Scholar]
- (84).Corcoran MM; Phad GE; Bernat NV; Stahl-Hennig C; Sumida N; Persson MAA; Martin M; Hedestam GBK Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat. Commun. 2016, 7, 13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


