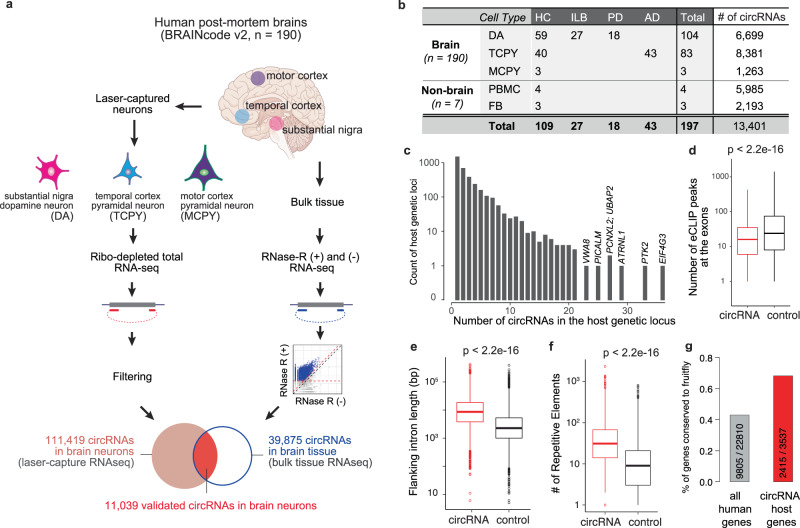
Fig. 1. Identification of circular RNAs actively transcribed in human brain neurons.
a Schema of circRNA identification in laser-captured brain neurons. In total, for 190 human post-mortem brains we performed total RNAseq for three types of neurons that were laser-captured from different brain regions and computationally identified 111,419 circRNAs. Experimental validation with paired RNase R- treated and untreated bulk tissue RNAseq was used to confirm 11,039 enriched circRNAs as the final set of validated circRNAs used for downstream analyses in this study. The brain image in panel a is credited to Анна Богатырева / Adobe Stock - stock.adobe.com. b Overview of samples and expressed circRNAs in each cell type. c Number of circRNAs per host genetic locus. d There are significantly less ENCODE-derived RNA-binding protein (RBP) eCLIP peaks overlapping with circRNAs (n = 10,845) vs. non-circularized exons (control, n = 10,845; two-sided Wilcox test, P < 2.2e−16). e Exons being circularized have longer flanking introns than background controls (n = 10,845; two-sided Wilcox test, P < 2.2e−16). f The flanking introns of circularized exons contain more repetitive elements than introns of control exons (n = 10,845; two-sided Wilcox test, P < 2.2e−16). g Proportions and numbers of human circRNA host genes conserved in Drosophila melanogaster. Grey bar shows the ratio of circRNA host genes to all genes in humans. Red bar shows the ratio of circRNA host genes to all genes in fruit fly.