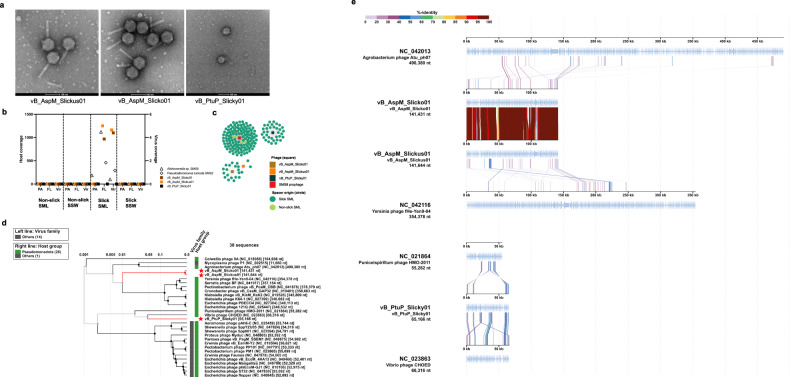
Fig. 8. Transmission electron microscopy images, abundance, CRISPR spacer matches and synteny of lytic phage isolates extracted from slick SML.
Negatively stained electron micrographs reveal myovirus morphology of Slickus and Slicko and podovirus morphology of Slicky, scale bar 100 nm (a). Coverage of reads based on mapping to the host MAGs Alishewanella sp. (MAG_01) and Pseudoalteromonas tunicata (MAG_66) as well as phages vB_AspM_Slickus01, vB_AspM_Slicko01, and vB_PtuP_Slicky01 (b). CRISPR spacer extracted from reads matching at 100% similarity to phage genomes from isolates and the SMS8 prophage of 50 kb. No matches from SSW spacers were detected. c Proteomic tree with placement of Slickus, Slicko, and Slicky (indicated by red stars) and closest related phages from the Refseq database (d). Based on insights from the tree, genomic alignments with related phages with indicated identity of proteins based on tBLASTX results (e). Annotations for the phage isolates are given in Table S14. SML sea-surface microlayer, SSW subsurface water.